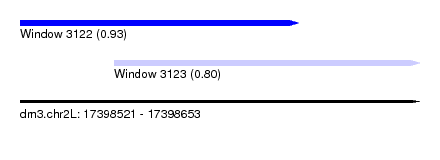
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,398,521 – 17,398,653 |
| Length | 132 |
| Max. P | 0.931682 |

| Location | 17,398,521 – 17,398,613 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 76.12 |
| Shannon entropy | 0.41929 |
| G+C content | 0.37855 |
| Mean single sequence MFE | -19.28 |
| Consensus MFE | -12.77 |
| Energy contribution | -12.80 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931682 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
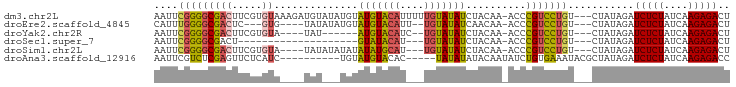
>dm3.chr2L 17398521 92 + 23011544 AAUUCGGGGCGACUUCGUGUAAAGAUGUAUAUGUAUGUACAUUUUUGUAUAUCUACAA-ACCCGUCCUGU---CUAUAGAUCUCUAUCAAGAGACU ....(((((((..((.(((((((((((((((....))))))))))))......))).)-)..))))))).---.......(((((....))))).. ( -22.70, z-score = -1.96, R) >droEre2.scaffold_4845 15736627 83 - 22589142 CAUUUGGGGCGACUC---GUG----UAUAUAUGUAUGUACAUU--UGUAUAUCAACAA-ACCCGUCCUGU---CUAUAGAUCUCUAUCAAGAGACU .....((((((....---(((----(((((....))))))))(--(((......))))-...))))))((---((((((....))))....)))). ( -19.40, z-score = -1.33, R) >droYak2.chr2R 17434113 80 - 21139217 AAUUCGGGGCGACUUCGUGUA----UAU------AUGUACAUC--UGUAUAUCUACAA-ACCCGUCCUGU---CUAUAGAUCUCUAUCAAGAGACU ....(((((((......((((----...------((((((...--.)))))).)))).-...))))))).---.......(((((....))))).. ( -19.70, z-score = -1.89, R) >droSec1.super_7 1040283 69 + 3727775 AAUUCGGGGCGACU--------------------GUAUACAU---UGUAUAUCUACAA-ACCCGUCCUGU---CUAUAGAUCUCUAUCAAGAGACU ....(((((((..(--------------------(....))(---((((....)))))-...))))))).---.......(((((....))))).. ( -15.30, z-score = -1.34, R) >droSim1.chr2L 17101087 85 + 22036055 AAUUCGGGGCGACUUCGUGUA----UAUAUAUAUAUAUGCAU---UGUAUAUCUACAA-ACCCGUCCUGU---CUAUAGAUCUCUAUCAAGAGACU ....(((((((......((((----((((....))))))))(---((((....)))))-...))))))).---.......(((((....))))).. ( -22.10, z-score = -2.48, R) >droAna3.scaffold_12916 12705171 81 - 16180835 AAUUCGUCUCGAGUUCUCAUC----------UGUAUGUACAC-----UAUAUAUACAAUAUCUGUGAAAUACGCUAUAGAUCUCUAUCAAGAGACC .....(((((...........----------((((((((...-----..))))))))..(((((((........))))))).........))))). ( -16.50, z-score = -1.64, R) >consensus AAUUCGGGGCGACUUCGUGUA____UAU___UGUAUGUACAU___UGUAUAUCUACAA_ACCCGUCCUGU___CUAUAGAUCUCUAUCAAGAGACU ....(((((((((.....))..............(((((((....)))))))..........)))))))...........(((((....))))).. (-12.77 = -12.80 + 0.03)
| Location | 17,398,552 – 17,398,653 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 77.59 |
| Shannon entropy | 0.42668 |
| G+C content | 0.45314 |
| Mean single sequence MFE | -17.75 |
| Consensus MFE | -9.80 |
| Energy contribution | -9.50 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802884 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
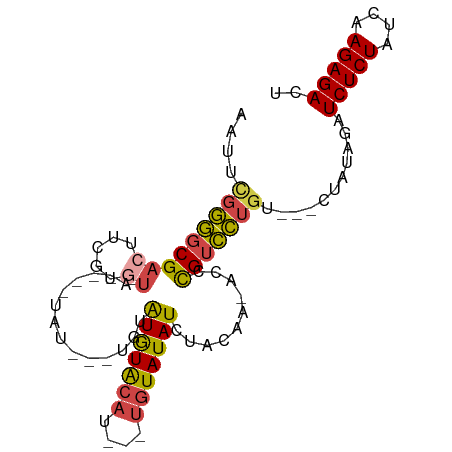
>dm3.chr2L 17398552 101 + 23011544 UGUAUGUACAUUUUUGUAUAUCUACAA-ACCCGUCCUGU---CUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUUCGCUCUCACCCACACCCG ((((((((((....))))))..)))).-...((.((((.---..((((.(((((....)))))))))))))))......(((.....)))............... ( -22.70, z-score = -2.91, R) >droEre2.scaffold_4845 15736651 99 - 22589142 UGUAUGUACAUUU--GUAUAUCAACAA-ACCCGUCCUGU---CUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUCACGCGAUCUUCACUCUCACCCACAUCCG ((.((((((....--))))))))....-......((((.---..((((.(((((....)))))))))))))(((.....)))....................... ( -18.20, z-score = -1.48, R) >droYak2.chr2R 17434136 97 - 21139217 --UAUGUACAUCU--GUAUAUCUACAA-ACCCGUCCUGU---CUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUUCACUCUCACCCACAUCCG --.((((((....--))))))......-......((((.---..((((.(((((....)))))))))))))(((.....)))....................... ( -17.00, z-score = -1.41, R) >droSec1.super_7 1040296 96 + 3727775 --UGUAUACAUU---GUAUAUCUACAA-ACCCGUCCUGU---CUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUUCGCUCUCACCCACAUCCG --.(((....((---(((....)))))-...((.((((.---..((((.(((((....)))))))))))))))...)))(((.....)))............... ( -19.80, z-score = -2.15, R) >droSim1.chr2L 17101114 98 + 22036055 UAUAUAUGCAUU---GUAUAUCUACAA-ACCCGUCCUGU---CUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUUCGCUCUCACCCACAUCCG .......((.((---(((....)))))-....(.((((.---..((((.(((((....)))))))))))))))).....(((.....)))............... ( -19.80, z-score = -2.10, R) >droWil1.scaffold_180772 883309 78 - 8906247 ------------------UAUAUGUAA-AU---UACAAU---UUA--GAUCUCUAUCAAGAGACUGUAAAGCGCUUUACGCGAUCCUCGCUCUCACCCACAUCCG ------------------.....((((-(.---......---.((--(.(((((....))))))))........)))))(((.....)))............... ( -12.49, z-score = -1.77, R) >droAna3.scaffold_12916 12705202 90 - 16180835 ---------------CUAUAUAUACAAUAUCUGUGAAAUACGCUAUAGAUCUCUAUCAAGAGACCGUCAGACGCUUCACGCGCUCCUCGCUCUCACCCACAUCCG ---------------................((((.........((((....))))...((((.((..((.(((.....)))))...)).))))...)))).... ( -15.30, z-score = -1.45, R) >droVir3.scaffold_13246 2084666 88 - 2672878 ----------------UCUAUAUAGUACUUAUAUCCGAUAC-UCAUAGAUCUCUAUCAAGAGACUGUCAAGCGCUUUACGCGCUCCUCGCUCUCGCCCUCGUCCG ----------------(((((..(((((........).)))-).)))))..........((((.((...(((((.....)))))...)).))))........... ( -16.70, z-score = -1.01, R) >consensus __UAU_UACAUU___GUAUAUCUACAA_ACCCGUCCUGU___CUAUAGAUCUCUAUCAAGAGACUGUCAGGCGCUUUACGCGAUCUUCGCUCUCACCCACAUCCG ...........................................................((((.((.....(((.....))).....)).))))........... ( -9.80 = -9.50 + -0.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:47:59 2011