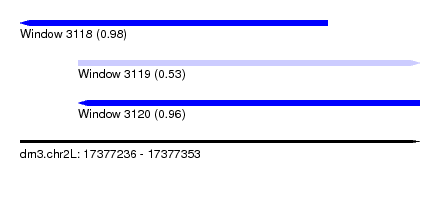
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,377,236 – 17,377,353 |
| Length | 117 |
| Max. P | 0.979653 |

| Location | 17,377,236 – 17,377,326 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 72.05 |
| Shannon entropy | 0.53610 |
| G+C content | 0.47010 |
| Mean single sequence MFE | -18.04 |
| Consensus MFE | -10.01 |
| Energy contribution | -9.98 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17377236 90 - 23011544 GCAAUUGCUUGAAUCCCCAU-UACCUCCAAGGAGCGUCAUAACCGAGACGUUCCCUGCUCCACUUACAUCAUCAAAUUGC---UCCAUUGCCAU ((((((...(((........-......((.((((((((........)))))))).))...........)))...))))))---........... ( -19.51, z-score = -3.35, R) >droSim1.chr2L 17079512 93 - 22036055 GCAAUUGUUUGAAUGCCCAU-UACCUCCAAGGAGCGUCAUAACCGAGACGUUCCCUGCUCCACUUACAUCAUCAAAUUGCUCCUCCAUUGCCAU ((((((...(((.((.....-......((.((((((((........)))))))).)).........)))))...)))))).............. ( -20.35, z-score = -3.07, R) >droSec1.super_7 1015297 93 - 3727775 GCAAUUGUUUGAAUGCCCAU-UACCUCCAAGGAGCGUCAUAACCGAGACGUUCCCGGCUCCACUUACAUCAUCAAAUUGCUCCUCCAUUGCCAU ((((((...(((.((.....-.....((..((((((((........)))))))).)).........)))))...)))))).............. ( -20.91, z-score = -2.56, R) >droYak2.chr2R 17412838 87 + 21139217 GCAAUUGUUUGAAUAUCUAU-UACCUCCAAGGAGCGUCAUAACCGAGACGUUCCCUGCUCCACUAACUUCA--AAUUG----UUCCACGGCUGU (.(((.(((((((.......-......((.((((((((........)))))))).))..........))))--))).)----)).)........ ( -16.90, z-score = -1.23, R) >droEre2.scaffold_4845 15713012 89 + 22589142 GUAAUGGCUUGGAUCUCCAU-UACCUCCAAGGAGCGUCAUAACCGAGACGUUCCCUGCUCCACUUACAUCAUCAAAUG----UUCCACGGCUGU ...((((((((((.......-......((.((((((((........)))))))).))........((((......)))----))))).)))))) ( -22.20, z-score = -1.73, R) >droPer1.super_1 4934900 88 - 10282868 -----GGAUUGUGCACUAAUAUACUUCCCAUAGAUGCCAUAUCCGUCUCUCUUGUAGUCCCCUUCUCAUCCACGG-UGUUUUUCUCAUUUCCCU -----((((((.((((((............))).))))).))))......................(((.....)-))................ ( -8.40, z-score = 0.50, R) >consensus GCAAUUGUUUGAAUACCCAU_UACCUCCAAGGAGCGUCAUAACCGAGACGUUCCCUGCUCCACUUACAUCAUCAAAUGGC__UUCCAUUGCCAU ..............................((((((((........))))))))........................................ (-10.01 = -9.98 + -0.02)



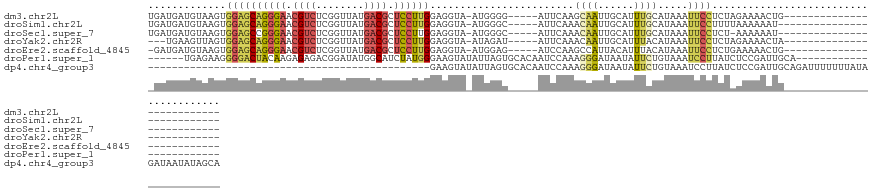
| Location | 17,377,253 – 17,377,353 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 132 |
| Reading direction | forward |
| Mean pairwise identity | 56.95 |
| Shannon entropy | 0.72677 |
| G+C content | 0.39488 |
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -8.23 |
| Energy contribution | -9.23 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.56 |
| Mean z-score | -1.10 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.531246 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17377253 100 + 23011544 UGAUGAUGUAAGUGGAGCAGGGAACGUCUCGGUUAUGACGCUCCUUGGAGGUA-AUGGGG-----AUUCAAGCAAUUGCAUUUGCAUAAAUUCCUCUAGAAAACUG-------------------------- ..........((((((((.(....)(((........))))))))(((((((..-(((.((-----((.(((....))).)))).))).....)))))))...))).-------------------------- ( -27.90, z-score = -1.47, R) >droSim1.chr2L 17079532 99 + 22036055 UGAUGAUGUAAGUGGAGCAGGGAACGUCUCGGUUAUGACGCUCCUUGGAGGUA-AUGGGC-----AUUCAAACAAUUGCAUUUGCAUAAAUUCCUUUUAAAAAAU--------------------------- .....(((((((((.((((((((.((((........)))).))))))(((((.-....))-----.)))......)).)))))))))..................--------------------------- ( -24.40, z-score = -1.20, R) >droSec1.super_7 1015317 98 + 3727775 UGAUGAUGUAAGUGGAGCCGGGAACGUCUCGGUUAUGACGCUCCUUGGAGGUA-AUGGGC-----AUUCAAACAAUUGCAUUUGCAUAAAUUCCUCU-AAAAAAU--------------------------- ..........((((.(((((((.....)))))))....))))..(((((((.(-((..((-----(..(((....)))....)))....))))))))-)).....--------------------------- ( -24.50, z-score = -0.91, R) >droYak2.chr2R 17412855 97 - 21139217 ---UGAAGUUAGUGGAGCAGGGAACGUCUCGGUUAUGACGCUCCUUGGAGGUA-AUAGAU-----AUUCAAACAAUUGCAUUUACAUAAAUUCCUCUAGAAAACUA-------------------------- ---.....((((.((((((((((.((((........)))).))))))...(((-((....-----.........)))))...........)))).)))).......-------------------------- ( -22.02, z-score = -1.08, R) >droEre2.scaffold_4845 15713029 99 - 22589142 -GAUGAUGUAAGUGGAGCAGGGAACGUCUCGGUUAUGACGCUCCUUGGAGGUA-AUGGAG-----AUCCAAGCCAUUACAUUUACAUAAAUUCCUCUGAAAAACUG-------------------------- -....(((((((((...((((((.((((........)))).))))))..(((.-.(((..-----..))).)))....)))))))))...................-------------------------- ( -29.50, z-score = -2.36, R) >droPer1.super_1 4934925 102 + 10282868 ------UGAGAAGGGGACUACAAGAGAGACGGAUAUGGCAUCUAUGGGAAGUAUAUUAGUGCACAAUCCAAAGGGAUAAUAUUCUGUAAAUCCUUAUCUCCGAUUGCA------------------------ ------.((((..((((.(((.........((((.((((((..(((.......)))..)))).))))))....(((......))))))..))))..))))........------------------------ ( -22.10, z-score = -0.06, R) >dp4.chr4_group3 3457389 85 + 11692001 -----------------------------------------------GAAGUAUAUUAGUGCACAAUCCAAAGGGAUAAUAUUCUGUAAAUCCUUAUCUCCGAUUGCAGAUUUUUUUAUAGAUAAUAUAGCA -----------------------------------------------....(((((((.......((((....)))).....((((((((.....((((........))))...)))))))))))))))... ( -13.70, z-score = -0.60, R) >consensus _GAUGAUGUAAGUGGAGCAGGGAACGUCUCGGUUAUGACGCUCCUUGGAGGUA_AUGGGU_____AUUCAAACAAUUGCAUUUGCAUAAAUUCCUCUAAAAAACUG__________________________ .............((((((((((.((((........)))).))))))........................((((......)))).....))))...................................... ( -8.23 = -9.23 + 1.00)


| Location | 17,377,253 – 17,377,353 |
|---|---|
| Length | 100 |
| Sequences | 7 |
| Columns | 132 |
| Reading direction | reverse |
| Mean pairwise identity | 56.95 |
| Shannon entropy | 0.72677 |
| G+C content | 0.39488 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -8.98 |
| Energy contribution | -8.51 |
| Covariance contribution | -0.46 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958807 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17377253 100 - 23011544 --------------------------CAGUUUUCUAGAGGAAUUUAUGCAAAUGCAAUUGCUUGAAU-----CCCCAU-UACCUCCAAGGAGCGUCAUAACCGAGACGUUCCCUGCUCCACUUACAUCAUCA --------------------------(((.......(.((.(((((.((((......)))).)))))-----.)))..-.........((((((((........)))))))))))................. ( -21.70, z-score = -2.02, R) >droSim1.chr2L 17079532 99 - 22036055 ---------------------------AUUUUUUAAAAGGAAUUUAUGCAAAUGCAAUUGUUUGAAU-----GCCCAU-UACCUCCAAGGAGCGUCAUAACCGAGACGUUCCCUGCUCCACUUACAUCAUCA ---------------------------...........((((((((.((((......)))).)))))-----......-......((.((((((((........)))))))).)).)))............. ( -20.30, z-score = -2.01, R) >droSec1.super_7 1015317 98 - 3727775 ---------------------------AUUUUUU-AGAGGAAUUUAUGCAAAUGCAAUUGUUUGAAU-----GCCCAU-UACCUCCAAGGAGCGUCAUAACCGAGACGUUCCCGGCUCCACUUACAUCAUCA ---------------------------.......-...((((((((.((((......)))).)))))-----(((...-.........((((((((........)))))))).))))))............. ( -20.72, z-score = -1.33, R) >droYak2.chr2R 17412855 97 + 21139217 --------------------------UAGUUUUCUAGAGGAAUUUAUGUAAAUGCAAUUGUUUGAAU-----AUCUAU-UACCUCCAAGGAGCGUCAUAACCGAGACGUUCCCUGCUCCACUAACUUCA--- --------------------------((((......(((((((..((((....((....))....))-----))..))-).))))((.((((((((........)))))))).))....))))......--- ( -21.60, z-score = -2.15, R) >droEre2.scaffold_4845 15713029 99 + 22589142 --------------------------CAGUUUUUCAGAGGAAUUUAUGUAAAUGUAAUGGCUUGGAU-----CUCCAU-UACCUCCAAGGAGCGUCAUAACCGAGACGUUCCCUGCUCCACUUACAUCAUC- --------------------------............(((............(((((((.......-----..))))-)))...((.((((((((........)))))))).)).)))............- ( -22.60, z-score = -1.15, R) >droPer1.super_1 4934925 102 - 10282868 ------------------------UGCAAUCGGAGAUAAGGAUUUACAGAAUAUUAUCCCUUUGGAUUGUGCACUAAUAUACUUCCCAUAGAUGCCAUAUCCGUCUCUCUUGUAGUCCCCUUCUCA------ ------------------------(((((..((((((..((((............))))....((((((.((((((............))).))))).)))))))))).)))))............------ ( -19.70, z-score = -0.59, R) >dp4.chr4_group3 3457389 85 - 11692001 UGCUAUAUUAUCUAUAAAAAAAUCUGCAAUCGGAGAUAAGGAUUUACAGAAUAUUAUCCCUUUGGAUUGUGCACUAAUAUACUUC----------------------------------------------- (((((((.....)))).........((((((((.(((((..((((...)))).)))))))....)))))))))............----------------------------------------------- ( -10.40, z-score = 0.41, R) >consensus __________________________CAGUUUUUGAGAGGAAUUUAUGCAAAUGCAAUUGUUUGAAU_____ACCCAU_UACCUCCAAGGAGCGUCAUAACCGAGACGUUCCCUGCUCCACUUACAUCAUC_ ......................................((((((((.((..........)).))))).....................((((((((........))))))))....)))............. ( -8.98 = -8.51 + -0.46)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:47:57 2011