
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,357,165 – 17,357,270 |
| Length | 105 |
| Max. P | 0.625769 |

| Location | 17,357,165 – 17,357,270 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.40 |
| Shannon entropy | 0.51888 |
| G+C content | 0.44027 |
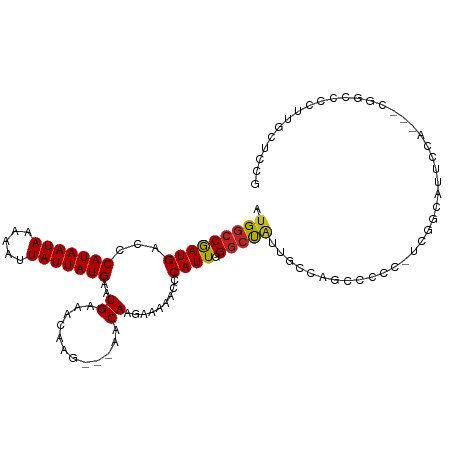
| Mean single sequence MFE | -20.10 |
| Consensus MFE | -9.68 |
| Energy contribution | -9.45 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
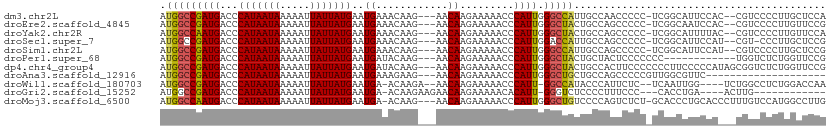
>dm3.chr2L 17357165 105 - 23011544 AUGGCCGAUGACCCAUAAUAAAAAUUAUUAUGAAUGAAACAAG---AACAAGAAAAACCCAUUGGGCCAUUGCCAACCCCC-UCGGCAUUCCAC--CGUCCCCUUGCUCCA ((((((.......(((((((.....)))))))...........---..(((..........)))))))))((((.......-..))))......--............... ( -17.40, z-score = -0.31, R) >droEre2.scaffold_4845 15693618 105 + 22589142 AUGGCCGAUGACCCAUAAUAAAAAUUAUUAUGAAUGAAACAAG---AACAAGAAAAACCCAUUGGGCUACUGCCAGCCCCC-UCGGCAAUCCAC--CGUCCCCUUGUUCCG .............(((((((.....)))))))..........(---((((((...........(((((......)))))..-.(((.......)--))....))))))).. ( -21.70, z-score = -2.06, R) >droYak2.chr2R 17392887 105 + 21139217 AUGGCCAAUGACCCAUAAUAAAAAUUAUUAUGAAUGAAACAAG---AACAAGAAAAACCCAUUGGGCUACUGCCAGCCCCC-UCGGCAUUUUAC--CGUCCCCUUGUUCCA .............(((((((.....)))))))..........(---((((((...........(((((......)))))..-.(((.......)--))....))))))).. ( -21.70, z-score = -1.82, R) >droSec1.super_7 995235 104 - 3727775 AUGGCCGAUGACCCAUAAUAAAAAUUAUUAUGAAUGAAACAAG---AACAAGAAAAACCCAUUGGACCAUUGCCAGCCCCC-UCGGCAUUCCAU--CGU-CCCUUGCUCCG ..((.(((((...(((((((.....)))))))...........---.....(((........(((.......)))(((...-..))).))))))--)).-))......... ( -17.90, z-score = -1.06, R) >droSim1.chr2L 17054592 105 - 22036055 AUGGCCGAUGACCCAUAAUAAAAAUUAUUAUGAAUGAAACAAG---AACAAGAAAAACCCAUUGGGCCAUUGCCAGCCCCC-UCGGCAUUCCAU--CGUCCCCUUGCUCCG ..((.(((((...(((((((.....)))))))...........---.....(((..........(((....))).(((...-..))).))))))--)).)).......... ( -20.20, z-score = -0.96, R) >droPer1.super_68 192360 96 + 338515 AUGGCCGAUGACCCAUAAUAAAAAUUAUUAUGAAUGAUACAAG---AACAAGAAAAACCCAUUGGGCUACUGCUACUCCCCCCC------------UGGUCUCUGGUUCCG ..(((((..(((((((((((.....)))))))...........---..................(((....)))..........------------.))))..)))))... ( -17.00, z-score = -0.46, R) >dp4.chr4_group4 295448 108 - 6586962 AUGGCCGAUGACCCAUAAUAAAAAUUAUUAUGAAUGAUACAAG---AACAAGAAAAACCCAUUGGGCUACUGCCACUUCCCCCCCUUCCCCCAUAGCGGUCUCUGGUUCCG ..(((((..(((((((((((.....)))))))...........---..................(((....))).......................))))..)))))... ( -19.60, z-score = -0.51, R) >droAna3.scaffold_12916 12665377 88 + 16180835 AUGGCCGAUGACCCAUAAUAAAAAUUAUUAUGAAUGAAAGAAG---AACAAGAAAAACCCAUUGGGCUGCUGCCAGCCCCCGUUGGCGUUC-------------------- ...(((((((...(((((((.....)))))))...........---.................((((((....)))))).)))))))....-------------------- ( -24.70, z-score = -3.29, R) >droWil1.scaffold_180703 3211171 101 + 3946847 AUGGCCGAUGACCCAUAAUAAAAAUUAUUAUGAAUGA-ACAAGA--AACAAGAAAAACCCAUU-GGCCAUACCCAUUCUC--UCAAUUGG----UCUGGCCUCUGGACCAA ((((((((((...(((((((.....)))))))..((.-......--..)).........))))-))))))..........--....((((----((..(...)..)))))) ( -26.60, z-score = -3.31, R) >droGri2.scaffold_15252 592358 90 + 17193109 AUGGCCGAUGACCCAUAAUAAAAAUUAUUAUGAAUGA-ACAAGAAGAACAAGAAAAACACAUU-GGGUCUCCCCUUUCCC---CACCUGA----ACUUG------------ ..((..((.(((((((((((.....)))))).((((.-.....................))))-))))))).))......---.......----.....------------ ( -10.35, z-score = 0.30, R) >droMoj3.scaffold_6500 5341178 106 - 32352404 AUGGCCAAUGACCCAUAAUAAAAAUUAUUAUGAAUGA-ACAAG---AACAAGAAAAACCCAUUGGGCUGUCCCCAGUCUCU-GCACCCUGCACCCUUUGUCCAUGGCCUUG ..((((.......(((((((.....))))))).(((.-(((((---..............((((((.....))))))...(-((.....)))...))))).)))))))... ( -23.90, z-score = -1.84, R) >consensus AUGGCCGAUGACCCAUAAUAAAAAUUAUUAUGAAUGAAACAAG___AACAAGAAAAACCCAUUGGGCUAUUGCCAGCCCCC_UCGGCAUUCCA___CGGCCCCUUGCUCCG .(((((((((...(((((((.....)))))))..((............)).........)))).))))).......................................... ( -9.68 = -9.45 + -0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:47:54 2011