| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,350,587 – 17,350,689 |
| Length | 102 |
| Max. P | 0.944870 |

| Location | 17,350,587 – 17,350,689 |
|---|---|
| Length | 102 |
| Sequences | 7 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.37 |
| Shannon entropy | 0.30340 |
| G+C content | 0.52028 |
| Mean single sequence MFE | -38.51 |
| Consensus MFE | -25.48 |
| Energy contribution | -25.19 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944870 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
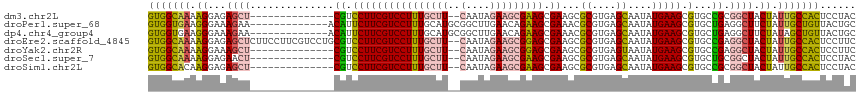
>dm3.chr2L 17350587 102 - 23011544 GUGGCAAAAGGAGAGCU--------------CGUCCUUCGUCCUUUGCUU--CAAUAGAAGCGAAGCGAAGCGCGUGAGCAAUAUGAAGCGUGCCGCGGCUACUAUUGCCACUCCUAC (((((((.((...(((.--------------(((..((((..((((((((--(....)))))))))))))((((((...(.....)..)))))).)))))).)).)))))))...... ( -41.90, z-score = -3.44, R) >droPer1.super_68 186059 105 + 338515 GUGGUGAAGGGAAAGAA-------------ACAUUCUUCGUCCUUUGCAUGCGGCUUGAACAGAAGCGAAACGCGUGAGCAAUAUGAAGCGUGCUGAGGCUUCUAUUGCUGUUACUGC (..(((((((((((((.-------------....))))..)))))).((((((..(((........)))..))))))(((((((.(((((.(....).))))))))))))..)))..) ( -34.40, z-score = -1.67, R) >dp4.chr4_group4 289163 105 - 6586962 GUGGUGAAGGGAAAGAA-------------ACAUUCUUCGUCCUUUGCAUGCGGCUUGAACAGAAGCGAAACGCGUGAGCAAUAUGAAGCGUGCUGAGGCUUCUAUAGCUGUUACUGC (..(((((((((((((.-------------....))))..))))))((((((.((((......)))).....((....))........))))))...((((.....))))..)))..) ( -31.20, z-score = -0.65, R) >droEre2.scaffold_4845 15685698 116 + 22589142 GUGGCAAAAGGAGAGCUCUUCCUUCGUCCUGCGUCCUUCGUCCUUUGCUU--CAAUAGAAGCGGAGCGAAGCGCGUGAGCAAUAUGAAGCGUGCCGAGGCUACUAUUGCCACUCCUUC (((((((((((((.....)))))).((((.(((..(((((((((((((((--(....))))))))).))...((....))....)))))..))).).))).....)))))))...... ( -44.20, z-score = -2.44, R) >droYak2.chr2R 17385962 102 + 21139217 GUGGCAAAAGGAAAGCU--------------CGUCCUUCGUCCUUUGCUU--CAAUAGAAGCGGAGCGAAGCGCGUGAGUAAUAUGAAGCGUGCCGAGGCUACUAUUGCCACUCCUUC (((((((.((...((((--------------((...((((..((((((((--(....)))))))))))))((((((............))))))))).))).)).)))))))...... ( -38.80, z-score = -2.93, R) >droSec1.super_7 988738 102 - 3727775 GUGGCAAAAGGAGAACU--------------CGUCCUUCGUCCUUUGCUU--CAAUAGAAGCGAAGCGAAGCGCGUGAGCAAUAUGAAGCGUGCUGCGGCUACUAUUGCCACUCCUAC (((((((((((((....--------------).))))).(((((((((((--(....))))))))).(.(((((((...(.....)..))))))).)))).....)))))))...... ( -38.30, z-score = -2.87, R) >droSim1.chr2L 17048850 102 - 22036055 GUGGCACAAGGAGAGCU--------------CGUCCUUCGUCCUUUGCUU--CAAUAGAAGCGAAGCGAAGCGCGUGAGCAAUAUGAAGCGUGCCGCGGCUACUAUUGCCACUCCUAC (((((((........(.--------------(((.(((((..((((((((--(....)))))))))))))).))).).((........)))))))))(((.......)))........ ( -40.80, z-score = -2.92, R) >consensus GUGGCAAAAGGAGAGCU______________CGUCCUUCGUCCUUUGCUU__CAAUAGAAGCGAAGCGAAGCGCGUGAGCAAUAUGAAGCGUGCCGAGGCUACUAUUGCCACUCCUAC (((((((.((.........................(((((((((((((((........)))))))).))...((....))....)))))...((....))..)).)))))))...... (-25.48 = -25.19 + -0.30)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:47:52 2011