| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,348,356 – 17,348,452 |
| Length | 96 |
| Max. P | 0.938949 |

| Location | 17,348,356 – 17,348,452 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 73.90 |
| Shannon entropy | 0.50997 |
| G+C content | 0.46069 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -15.10 |
| Energy contribution | -14.99 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.938949 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
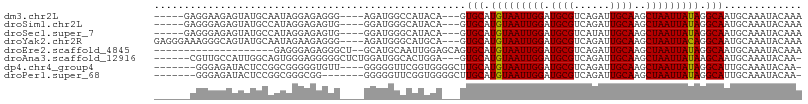
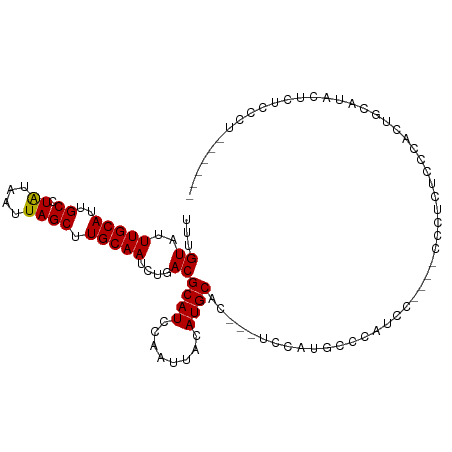
>dm3.chr2L 17348356 96 + 23011544 -----GAGGAAGAGUAUGCAAUAGGAGAGGG----AGAUGGCCAUACA---GUGCAUGUAAUUGGAUGCGUCAGAUUGCAAGCUAAUUAUAGGCAAUGCAAAUACAAA -----........(((((((.........((----......)).....---.(((.(((((((((.((((......))))..))))))))).))).))))..)))... ( -20.90, z-score = -1.20, R) >droSim1.chr2L 17046564 96 + 22036055 -----GAGGGAGAGUAUGCCAUAGGAGAGUG----GGAUGGGCAUACA---GUGCAUGUAAUUGGAUGCGUCAGAUUGCAAGCUAAUUAUAGGCAAUGCAAAUACAAA -----........(((((((...........----.....))))))).---.(((.(((((((((.((((......))))..))))))))).)))............. ( -24.49, z-score = -1.85, R) >droSec1.super_7 986463 96 + 3727775 -----GAGGGAGAGUAUGCCAUAGGAGAGUG----GGAUGGGCAUACA---GUGCAUGUAAUUGGAUGCGUCAUAUUGCAAGCUAAUUAUAGGCAAUGCAAAUACAAA -----........(((((((...........----.....))))))).---.(((.(((((((((.((((......))))..))))))))).)))............. ( -24.49, z-score = -1.69, R) >droYak2.chr2R 17382597 101 - 21139217 GAGGGAAAGGGCAGUAUGCAAUAGAAGAGGG----AGAUGGGCAUGCA---GUGCAUGUAAUUGGAUGCGUCAGAUUGCAAGCUAAUUACAGGCAAUGCAAAUACAAA ..........(((((((((............----......)))))).---.(((.(((((((((.((((......))))..))))))))).))).)))......... ( -26.17, z-score = -2.73, R) >droEre2.scaffold_4845 15683530 86 - 22589142 --------------------GAGGGAGAGGGCU--GCAUGCAAUUGGAGCAGUGCAUGUAAUUGGAUGCGUCAGAUUGCAAGCUAAUUAUAGGCAAUGCAAAUACAAA --------------------......((..((.--....))..))...(((.(((.(((((((((.((((......))))..))))))))).))).)))......... ( -21.80, z-score = -0.65, R) >droAna3.scaffold_12916 12658258 98 - 16180835 ------CGUUGCCAUUGGCAGUGGGAGGGGGCUCUGGAUGGCACUGGA---GUGCAUGUAAUUGGAUGCGUCAGAUUGCAAGCUAAUUAUAAGCAAUGCAAAUACAA- ------...(((((((.(.(((........))).).))))))).((.(---.(((.(((((((((.((((......))))..))))))))).))).).)).......- ( -27.60, z-score = -0.26, R) >dp4.chr4_group4 285709 96 + 6586962 -------GGGAGAUACUCCGGCGGGGGUGUU----GGGGGUUCGGUGGGGCUUGCAUGUAAUUGGAUGCGUCAGAUUGCAAGCUAAUUAUAGGCAUUGCAAAUACAA- -------....((..((((((((....))))----))))..)).(((..((.(((.(((((((((.((((......))))..))))))))).)))..))...)))..- ( -30.70, z-score = -2.30, R) >droPer1.super_68 182169 93 - 338515 -------GGGAGAUACUCCGGCGGGCGG-------GGGGGUUCGGUGGGGCUUGCAUGUAAUUGGAUGCGUCAGAUUGCAAGCUAAUUAUAGGCAUUGCAAAUACAA- -------..(((...(((((.....)))-------))...))).(((..((.(((.(((((((((.((((......))))..))))))))).)))..))...)))..- ( -29.10, z-score = -1.91, R) >consensus ______AGGGAGAGUAUGCAAUAGGAGAGGG____GGAUGGGCAUACA___GUGCAUGUAAUUGGAUGCGUCAGAUUGCAAGCUAAUUAUAGGCAAUGCAAAUACAAA ....................................................(((.(((((((((.((((......))))..))))))))).)))............. (-15.10 = -14.99 + -0.11)
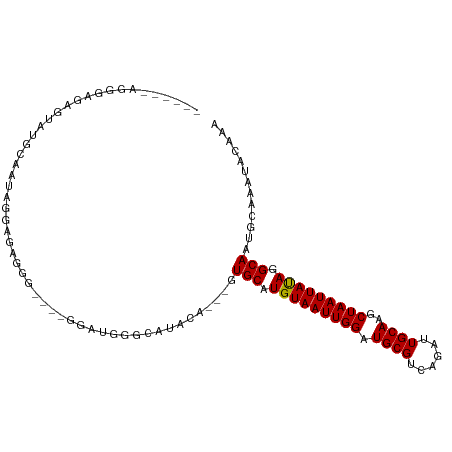
| Location | 17,348,356 – 17,348,452 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 73.90 |
| Shannon entropy | 0.50997 |
| G+C content | 0.46069 |
| Mean single sequence MFE | -15.79 |
| Consensus MFE | -8.77 |
| Energy contribution | -8.66 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.691930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17348356 96 - 23011544 UUUGUAUUUGCAUUGCCUAUAAUUAGCUUGCAAUCUGACGCAUCCAAUUACAUGCAC---UGUAUGGCCAUCU----CCCUCUCCUAUUGCAUACUCUUCCUC----- ...((..(((((..((.((....)))).)))))....))((((........))))..---.(((((.......----.............)))))........----- ( -11.45, z-score = 0.09, R) >droSim1.chr2L 17046564 96 - 22036055 UUUGUAUUUGCAUUGCCUAUAAUUAGCUUGCAAUCUGACGCAUCCAAUUACAUGCAC---UGUAUGCCCAUCC----CACUCUCCUAUGGCAUACUCUCCCUC----- ...((..(((((..((.((....)))).)))))....))((((........))))..---.(((((((.....----...........)))))))........----- ( -17.79, z-score = -2.29, R) >droSec1.super_7 986463 96 - 3727775 UUUGUAUUUGCAUUGCCUAUAAUUAGCUUGCAAUAUGACGCAUCCAAUUACAUGCAC---UGUAUGCCCAUCC----CACUCUCCUAUGGCAUACUCUCCCUC----- .(..((((.(((..((.((....)))).)))))))..).((((........))))..---.(((((((.....----...........)))))))........----- ( -17.79, z-score = -2.10, R) >droYak2.chr2R 17382597 101 + 21139217 UUUGUAUUUGCAUUGCCUGUAAUUAGCUUGCAAUCUGACGCAUCCAAUUACAUGCAC---UGCAUGCCCAUCU----CCCUCUUCUAUUGCAUACUGCCCUUUCCCUC ...(((..((((.(((.(((((((.(..(((........)))..)))))))).))).---)))))))......----............((.....)).......... ( -18.60, z-score = -2.61, R) >droEre2.scaffold_4845 15683530 86 + 22589142 UUUGUAUUUGCAUUGCCUAUAAUUAGCUUGCAAUCUGACGCAUCCAAUUACAUGCACUGCUCCAAUUGCAUGC--AGCCCUCUCCCUC-------------------- ...((..(((((..((.((....)))).)))))....))((((.(((((....((...))...))))).))))--.............-------------------- ( -13.80, z-score = -0.30, R) >droAna3.scaffold_12916 12658258 98 + 16180835 -UUGUAUUUGCAUUGCUUAUAAUUAGCUUGCAAUCUGACGCAUCCAAUUACAUGCAC---UCCAGUGCCAUCCAGAGCCCCCUCCCACUGCCAAUGGCAACG------ -..((..(((((..(((.......))).)))))....))((((........))))..---.....((((((.(((.(........).)))...))))))...------ ( -19.30, z-score = -0.96, R) >dp4.chr4_group4 285709 96 - 6586962 -UUGUAUUUGCAAUGCCUAUAAUUAGCUUGCAAUCUGACGCAUCCAAUUACAUGCAAGCCCCACCGAACCCCC----AACACCCCCGCCGGAGUAUCUCCC------- -..((..((((((.((.((....))))))))))....))((((........))))..................----............((((...)))).------- ( -13.80, z-score = -0.81, R) >droPer1.super_68 182169 93 + 338515 -UUGUAUUUGCAAUGCCUAUAAUUAGCUUGCAAUCUGACGCAUCCAAUUACAUGCAAGCCCCACCGAACCCCC-------CCGCCCGCCGGAGUAUCUCCC------- -..((..((((((.((.((....))))))))))....))((((........))))..................-------.........((((...)))).------- ( -13.80, z-score = -0.36, R) >consensus UUUGUAUUUGCAUUGCCUAUAAUUAGCUUGCAAUCUGACGCAUCCAAUUACAUGCAC___UCCAUGCCCAUCC____CCCUCUCCCACUGCAUACUCUCCCU______ ...((..(((((..((.((....)))).)))))....))((((........))))..................................................... ( -8.77 = -8.66 + -0.11)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:47:51 2011