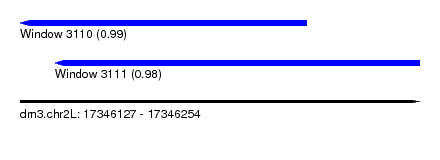
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,346,127 – 17,346,254 |
| Length | 127 |
| Max. P | 0.993026 |

| Location | 17,346,127 – 17,346,218 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 82.06 |
| Shannon entropy | 0.35834 |
| G+C content | 0.40375 |
| Mean single sequence MFE | -24.51 |
| Consensus MFE | -21.58 |
| Energy contribution | -21.45 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.58 |
| SVM RNA-class probability | 0.993026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
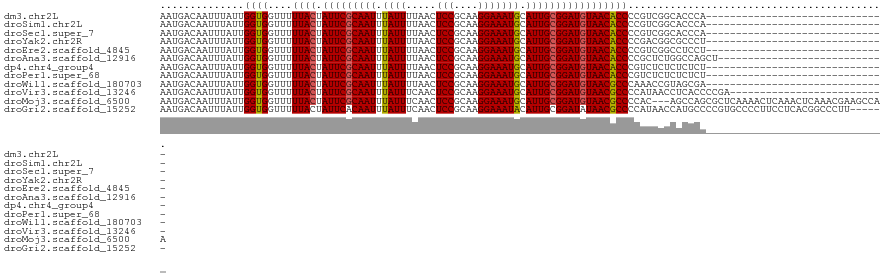
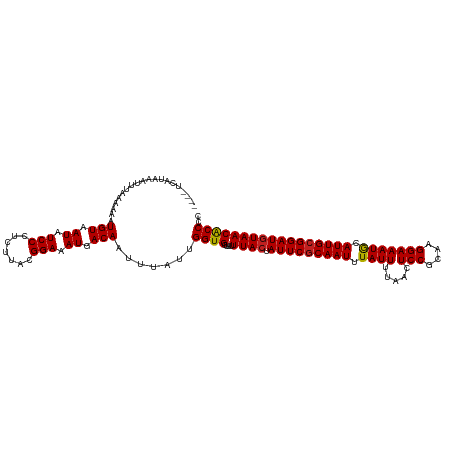
>dm3.chr2L 17346127 91 - 23011544 AAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCCGUCGGCACCCA------------------------------ ..((((........((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))))))))).......------------------------------ ( -24.60, z-score = -2.14, R) >droSim1.chr2L 17044362 91 - 22036055 AAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCCGUCGGCACCCA------------------------------ ..((((........((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))))))))).......------------------------------ ( -24.60, z-score = -2.14, R) >droSec1.super_7 978702 91 - 3727775 AAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCCGUCGGCACCCA------------------------------ ..((((........((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))))))))).......------------------------------ ( -24.60, z-score = -2.14, R) >droYak2.chr2R 17379256 91 + 21139217 AAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCCGACGGCGCCCU------------------------------ .........(..((((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).)))))))..)......------------------------------ ( -25.90, z-score = -2.52, R) >droEre2.scaffold_4845 15681431 91 + 22589142 AAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCCGUCGGCCUCCU------------------------------ ..((((........((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))))))))).......------------------------------ ( -24.60, z-score = -2.37, R) >droAna3.scaffold_12916 12656613 93 + 16180835 AAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCCGCUCUGGCCAGCU---------------------------- ..............((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).)))))(((......))).---------------------------- ( -26.10, z-score = -2.44, R) >dp4.chr4_group4 282632 91 - 6586962 AAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCGUCUCUCUCUCU------------------------------ ..............((((....((((.(((((((((.((((.....(((....))))))).))))))))))))))))).............------------------------------ ( -22.40, z-score = -3.23, R) >droPer1.super_68 178001 91 + 338515 AAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCGUCUCUCUCUCU------------------------------ ..............((((....((((.(((((((((.((((.....(((....))))))).))))))))))))))))).............------------------------------ ( -22.40, z-score = -3.23, R) >droWil1.scaffold_180703 3191632 91 + 3946847 AAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACGCCCAAACCGUAGCGA------------------------------ ..............((((((..((((.(((((((((.((((.....(((....))))))).))))))))))))).)))...))).......------------------------------ ( -23.70, z-score = -1.95, R) >droVir3.scaffold_13246 2026402 95 + 2672878 AAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUCAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACGCCCCAUAACCUCACCCCGA-------------------------- .............(((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))))))..............-------------------------- ( -25.40, z-score = -2.88, R) >droMoj3.scaffold_6500 5326811 118 - 32352404 AAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUCAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACGCCCCAC---AGCCAGCGCUCAAAACUCAAACUCAAACGAAGCCAA ..(((........(((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).)))))).---(((....)))......)))................. ( -26.60, z-score = -1.35, R) >droGri2.scaffold_15252 16584643 115 - 17193109 AAUGACAAUUUAUUGGUGGUUUUUACUAUUCACAAUUUAUUUCAACUCCGCAAGGAAAUACAUUGCGGAUAUAACGCCCCAUAACCAUGCCCCGUGCCCCUUCCUCACGGCCCUU------ .............(((.(((.((...(((((.((((.((((.....(((....))))))).)))).))))).)).))))))..........(((((.........))))).....------ ( -23.20, z-score = -1.17, R) >consensus AAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCCGUCGGCACCCA______________________________ ..............((((....((((.(((((((((.((((.....(((....))))))).)))))))))))))))))........................................... (-21.58 = -21.45 + -0.13)
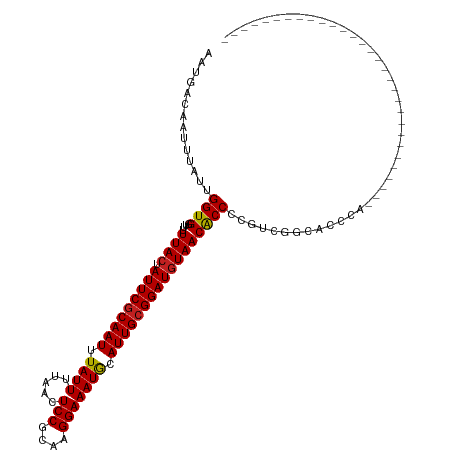
| Location | 17,346,138 – 17,346,254 |
|---|---|
| Length | 116 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.86 |
| Shannon entropy | 0.04723 |
| G+C content | 0.33156 |
| Mean single sequence MFE | -25.66 |
| Consensus MFE | -24.88 |
| Energy contribution | -24.75 |
| Covariance contribution | -0.13 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.980640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17346138 116 - 23011544 ----UCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCC ----................(((.((.(((.......))).)).))).......((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))))) ( -26.00, z-score = -2.50, R) >droSim1.chr2L 17044373 116 - 22036055 ----UCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCC ----................(((.((.(((.......))).)).))).......((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))))) ( -26.00, z-score = -2.50, R) >droSec1.super_7 978713 116 - 3727775 ----UCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCC ----................(((.((.(((.......))).)).))).......((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))))) ( -26.00, z-score = -2.50, R) >droYak2.chr2R 17379267 116 + 21139217 ----UCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCC ----................(((.((.(((.......))).)).))).......((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))))) ( -26.00, z-score = -2.50, R) >droEre2.scaffold_4845 15681442 116 + 22589142 ----UCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCC ----................(((.((.(((.......))).)).))).......((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))))) ( -26.00, z-score = -2.50, R) >droAna3.scaffold_12916 12656626 116 + 16180835 ----UCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCC ----................(((.((.(((.......))).)).))).......((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))))) ( -26.00, z-score = -2.50, R) >dp4.chr4_group4 282643 116 - 6586962 ----UCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCG ----................(((.((.(((.......))).)).))).......((((....((((.(((((((((.((((.....(((....))))))).))))))))))))))))).. ( -25.70, z-score = -2.49, R) >droPer1.super_68 178012 116 + 338515 ----UCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCG ----................(((.((.(((.......))).)).))).......((((....((((.(((((((((.((((.....(((....))))))).))))))))))))))))).. ( -25.70, z-score = -2.49, R) >droWil1.scaffold_180703 3191643 116 + 3946847 ----UCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACGCCCA ----................(((.((.(((.......))).)).))).......((((....((((.(((((((((.((((.....(((....))))))).))))))))))))))))).. ( -25.30, z-score = -1.97, R) >droVir3.scaffold_13246 2026417 116 + 2672878 ----UCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUCAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACGCCCC ----................(((.((.(((.......))).)).))).......((.(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))))) ( -26.30, z-score = -2.27, R) >droMoj3.scaffold_6500 5326849 120 - 32352404 GCGCUCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUCAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACGCCCC ..(((.(((((((.......(((((........)))))........))))))).)))(((..((((.(((((((((.((((.....(((....))))))).))))))))))))).))).. ( -27.36, z-score = -1.93, R) >droGri2.scaffold_15252 16584678 116 - 17193109 ----UCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCACAAUUUAUUUCAACUCCGCAAGGAAAUACAUUGCGGAUAUAACGCCCC ----.................((.((((((.......(((..(((.(((..(((((((((....)))))...))))..))))))..)))(((((......).)))))))))).))..... ( -21.50, z-score = -1.69, R) >consensus ____UCAUAAAUUUAAAAAAUGUAAUAUCCCUCUUACGGAAAUGACAAUUUAUUGGUGGUUUUUACUAUUCGCAAUUUAUUUUAACUCCGCAAGGAAAUGCAUUGCGGAUGUAACACCCC ....................(((.((.(((.......))).)).))).......((((....((((.(((((((((.((((.....(((....))))))).))))))))))))))))).. (-24.88 = -24.75 + -0.13)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:47:50 2011