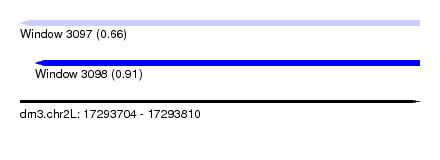
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,293,704 – 17,293,810 |
| Length | 106 |
| Max. P | 0.909380 |

| Location | 17,293,704 – 17,293,810 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.33 |
| Shannon entropy | 0.32500 |
| G+C content | 0.49492 |
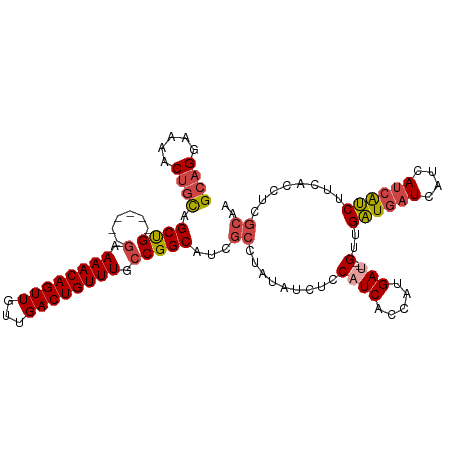
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -17.85 |
| Energy contribution | -17.72 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.662585 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
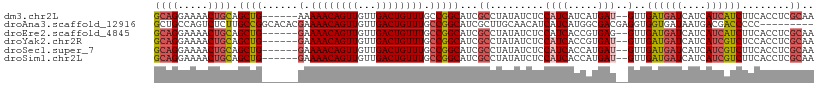
>dm3.chr2L 17293704 106 - 23011544 GCAGGAAAACUGCAGCUG------AAAAACAGUUGUUGACUGUUUGCCGGCAUCGCCUAUAUCUCCAUCAUCAUGAU--GUUGAUGAUCAUCAUCAUCUUCACCUCGCAACAGG ((((.....)))).((((------..((((((((...))))))))..))))...((.........((((.....)))--)..((((((....))))))........))...... ( -29.00, z-score = -1.72, R) >droAna3.scaffold_12916 12610928 101 + 16180835 GCUGCCAGUCUCUUGCCGGCACACGAAAACAGUUGUUGACUGUUUGCCGGCAUCGCUUGCAACAUCAUCAUGGCGACGAGGUGGUGAUAAUGACGACCCCC------------- ..(((.(((....((((((((......(((((((...)))))))))))))))..))).))).(((((((.((....)).)))))))...............------------- ( -35.10, z-score = -1.59, R) >droEre2.scaffold_4845 15631344 106 + 22589142 GCAGGAAAACUGCAGCUG------GAAAACAGUUGUUGACUGUUUGCCGGCAUCGCCUAUAUCUCCAUCACCGUGAG--GUUGAUGAUCAUCAUCAUCUUCACCUCGCAACAGG ((((.....)))).((((------(.((((((((...)))))))).))))).....................(((((--((.((((((....))))))...)))))))...... ( -39.40, z-score = -4.07, R) >droYak2.chr2R 17325845 106 + 21139217 GCAGGAAAACUGCAGCUG------GAAAACAGUUGUUGACUGUUUGCCGGCAUCGCCUAUAUCUCCAUCACCGUGAU--GUUGAUGAUCAUCAUCGUCUCCACCUCGCAACAGU ((((.....)))).((((------(.((((((((...)))))))).))))).....................((((.--((.((((((....))))))...)).))))...... ( -32.50, z-score = -2.15, R) >droSec1.super_7 926954 106 - 3727775 GCAGGAAAACUGCAGCUG------GAAAACAGUUGUUGACUGUUUGCCGGCAUCGCCUAUAUCUCCAUCACCAUGAU--GUUGAUGAUCAUCAUCGUCUUCACCUCGCAACGGG ((((.....)))).((((------(.((((((((...)))))))).)))))...((.........((((.....)))--)..((((((....))))))........))...... ( -31.60, z-score = -1.71, R) >droSim1.chr2L 16989193 106 - 22036055 GCAGGAAAACUGCAGCUG------GAAAACAGUUGUUGACUGUUUGCCGGCAUCGCCUAUAUCUCCAUCACCAUGAU--GUUGAUGAUCAUCAUCGUCUUCACCUCGCAACGGG ((((.....)))).((((------(.((((((((...)))))))).)))))...((.........((((.....)))--)..((((((....))))))........))...... ( -31.60, z-score = -1.71, R) >consensus GCAGGAAAACUGCAGCUG______GAAAACAGUUGUUGACUGUUUGCCGGCAUCGCCUAUAUCUCCAUCACCAUGAU__GUUGAUGAUCAUCAUCAUCUUCACCUCGCAACAGG ((((.....)))).((((........((((((((...))))))))..))))...............................((((((....))))))................ (-17.85 = -17.72 + -0.14)



| Location | 17,293,708 – 17,293,810 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.57 |
| Shannon entropy | 0.30036 |
| G+C content | 0.48457 |
| Mean single sequence MFE | -33.20 |
| Consensus MFE | -17.85 |
| Energy contribution | -17.72 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.909380 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17293708 102 - 23011544 GCAGGAAAACUGCAGCUG------AAAAACAGUUGUUGACUGUUUGCCGGCAUCGCCUAUAUCUCCAUCAUCAUGAU--GUUGAUGAUCAUCAUCAUCUUCACCUCGCAA ((((.....)))).((((------..((((((((...))))))))..))))...((.........((((.....)))--)..((((((....))))))........)).. ( -29.00, z-score = -2.27, R) >droAna3.scaffold_12916 12610928 101 + 16180835 GCUGCCAGUCUCUUGCCGGCACACGAAAACAGUUGUUGACUGUUUGCCGGCAUCGCUUGCAACAUCAUCAUGGCGACGAGGUGGUGAUAAUGACGACCCCC--------- ..(((.(((....((((((((......(((((((...)))))))))))))))..))).))).(((((((.((....)).)))))))...............--------- ( -35.10, z-score = -1.59, R) >droEre2.scaffold_4845 15631348 102 + 22589142 GCAGGAAAACUGCAGCUG------GAAAACAGUUGUUGACUGUUUGCCGGCAUCGCCUAUAUCUCCAUCACCGUGAG--GUUGAUGAUCAUCAUCAUCUUCACCUCGCAA ((((.....)))).((((------(.((((((((...)))))))).))))).....................(((((--((.((((((....))))))...))))))).. ( -39.40, z-score = -4.64, R) >droYak2.chr2R 17325849 102 + 21139217 GCAGGAAAACUGCAGCUG------GAAAACAGUUGUUGACUGUUUGCCGGCAUCGCCUAUAUCUCCAUCACCGUGAU--GUUGAUGAUCAUCAUCGUCUCCACCUCGCAA ((((.....)))).((((------(.((((((((...)))))))).))))).....................((((.--((.((((((....))))))...)).)))).. ( -32.50, z-score = -2.67, R) >droSec1.super_7 926958 102 - 3727775 GCAGGAAAACUGCAGCUG------GAAAACAGUUGUUGACUGUUUGCCGGCAUCGCCUAUAUCUCCAUCACCAUGAU--GUUGAUGAUCAUCAUCGUCUUCACCUCGCAA ((((.....)))).((((------(.((((((((...)))))))).)))))...((.........((((.....)))--)..((((((....))))))........)).. ( -31.60, z-score = -2.56, R) >droSim1.chr2L 16989197 102 - 22036055 GCAGGAAAACUGCAGCUG------GAAAACAGUUGUUGACUGUUUGCCGGCAUCGCCUAUAUCUCCAUCACCAUGAU--GUUGAUGAUCAUCAUCGUCUUCACCUCGCAA ((((.....)))).((((------(.((((((((...)))))))).)))))...((.........((((.....)))--)..((((((....))))))........)).. ( -31.60, z-score = -2.56, R) >consensus GCAGGAAAACUGCAGCUG______GAAAACAGUUGUUGACUGUUUGCCGGCAUCGCCUAUAUCUCCAUCACCAUGAU__GUUGAUGAUCAUCAUCAUCUUCACCUCGCAA ((((.....)))).((((........((((((((...))))))))..))))...............................((((((....))))))............ (-17.85 = -17.72 + -0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:47:39 2011