| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,143,601 – 17,143,698 |
| Length | 97 |
| Max. P | 0.678636 |

| Location | 17,143,601 – 17,143,698 |
|---|---|
| Length | 97 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 75.41 |
| Shannon entropy | 0.49311 |
| G+C content | 0.34652 |
| Mean single sequence MFE | -19.64 |
| Consensus MFE | -10.36 |
| Energy contribution | -11.41 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.678636 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
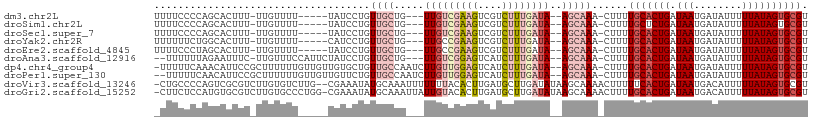
>dm3.chr2L 17143601 97 + 23011544 UUUUCCCCAGCACUUU-UUGUUUU-----UAUCCUGUUGCUG---UUGUCGAAGUCGUCUUUGAUA--AGCAAA-CUUUUGCACUGAUAAUGAUAUUUUUAUAGUGCGU .........(((((..-.((((.(-----((((..((.....---(((((((((....))))))))--)((((.-...)))))).))))).)))).......))))).. ( -21.90, z-score = -3.01, R) >droSim1.chr2L 16848978 97 + 22036055 UUUUCCCCAGCACUUU-UUGUUUU-----UAUCCUGUUGCUG---UUGUCGAAGUCGUCUUUGAUA--AGCAAA-CUUUUGCUCUGAUAAUGAUAUUUUUAUAGUGCGU .........(((((..-.((((.(-----((((.........---(((((((((....))))))))--)((((.-...))))...))))).)))).......))))).. ( -21.40, z-score = -3.18, R) >droSec1.super_7 779852 97 + 3727775 UUUUCCCCAGCACUUU-UUGUUUU-----UAUCCUGUUGCUG---UUGUCGAAGUCGUCUUUGAUA--AGCAAA-CUUUUGCACUGAUAAUGAUAUUUUUAUAGUGCGU .........(((((..-.((((.(-----((((..((.....---(((((((((....))))))))--)((((.-...)))))).))))).)))).......))))).. ( -21.90, z-score = -3.01, R) >droYak2.chr2R 17187732 97 - 21139217 UUUUUUCUGGCACUUU-UUGUUUU-----CAUCCUGUUGCUG---UUGCCGAAGUCGUCUUUGAUA--AGCAAA-CUUUUGCACUGAUAAUGAUAUUUUUAUAGUGCGU .........(((((..-......(-----(((.....(((..---(((((((((....)))))...--.)))).-.....)))......)))).........))))).. ( -15.73, z-score = 0.17, R) >droEre2.scaffold_4845 15496812 97 - 22589142 UUUUCCCUAGCACUUU-UUGUUUU-----UAUCCUGUUGCUG---UUGCCGAAGUCGUCUUUGAUA--AGCAAA-CUUUUGCACUGAUAAUGAUAUUUUUAUAGUGCGU .........(((((..-.((((.(-----((((..((.((..---(((((((((....)))))...--.)))).-.....)))).))))).)))).......))))).. ( -18.70, z-score = -1.77, R) >droAna3.scaffold_12916 12471786 100 - 16180835 --UUUUUUAGAAUUUC-UUGUUUCCAUUCUAUCCUGUUGCUG---UUGUCGGAGUCAUCUUUGAUA--AGCAAA-CUUUUGCACUGAUAAUGAUAUUUUUAUAGUGCGU --.....((((((...-........)))))).....(((((.---.((((((((....))))))))--))))).-.....((((((.(((........))))))))).. ( -20.60, z-score = -1.99, R) >dp4.chr4_group4 41772 105 + 6586962 -UUUUUCAAACAUUCCGCUUUUUUGUUGUUGUGCUGUUGCCAAUCUUGUUGGAGUCAUCUUUGAUA--AGCAAA-CUUUUGCACUGAUAAUGAUAUUUUUAUAGUGCGU -....................((((..((((.((....))))))(((((..(((....)))..)))--))))))-.....((((((.(((........))))))))).. ( -19.70, z-score = -0.41, R) >droPer1.super_130 44107 104 - 96376 --UUUUUCAACAUUCCGCUUUUUUGUUGUUGUUCUGUUGCCAAUCUUGUUGGAGUCAUCUUUGAUA--AGCAAA-CUUUUGCACUGAUAAUGAUAUUUUUAUAGUGCGU --.....(((((...........)))))................(((((..(((....)))..)))--))....-.....((((((.(((........))))))))).. ( -18.40, z-score = -0.49, R) >droVir3.scaffold_13246 1811560 106 - 2672878 -CUGCCCCAGUCGCGUCUUGUGUCUUG--CGAAAUAUGCAAAUUUUUUUACACUUGAUGCUUGAUAUAAGCAAAACUUUUUCACUGAUAAUGACAUUUUUAUAGUGCGU -.(((....((((((((..((((.(((--((.....)))))........))))..)))))..)))....))).........(((((.(((........))))))))... ( -17.80, z-score = 0.12, R) >droGri2.scaffold_15252 16360570 107 + 17193109 -CUUCUCCAUGUGCGUCUUGUGCCCUGG-CGAAAUAUGCAAAUUAUUGUACACUUGAUGCUUGAUAUAAGCAAAACUUUUGCACUGAUAAUGACAUUUUUAUAGUGCGU -......((...(((((..((((....)-)......(((((....))))).))..))))).)).................((((((.(((........))))))))).. ( -20.30, z-score = -0.01, R) >consensus _UUUCCCCAGCACUUU_UUGUUUU_____UAUCCUGUUGCUG___UUGUCGAAGUCAUCUUUGAUA__AGCAAA_CUUUUGCACUGAUAAUGAUAUUUUUAUAGUGCGU ....................................((((......((((((((....))))))))...))))......(((((((.(((........)))))))))). (-10.36 = -11.41 + 1.05)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:47:21 2011