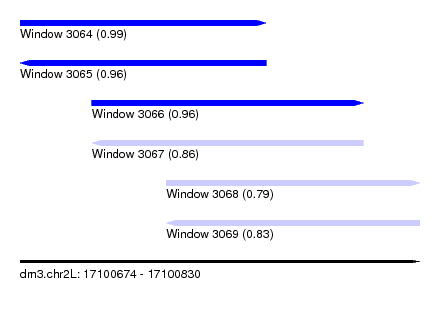
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,100,674 – 17,100,830 |
| Length | 156 |
| Max. P | 0.993612 |

| Location | 17,100,674 – 17,100,770 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 69.23 |
| Shannon entropy | 0.65789 |
| G+C content | 0.44027 |
| Mean single sequence MFE | -26.16 |
| Consensus MFE | -9.56 |
| Energy contribution | -9.12 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.63 |
| SVM RNA-class probability | 0.993612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
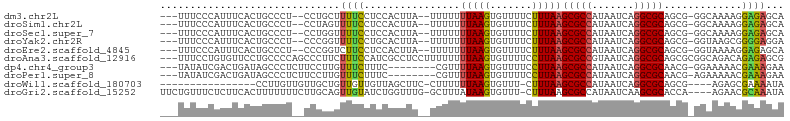
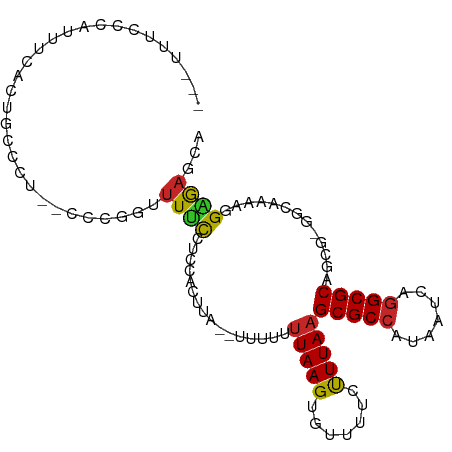
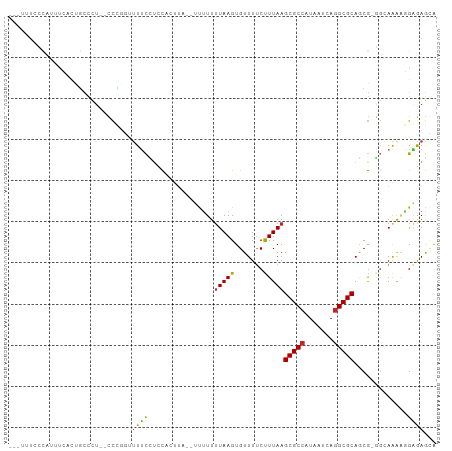
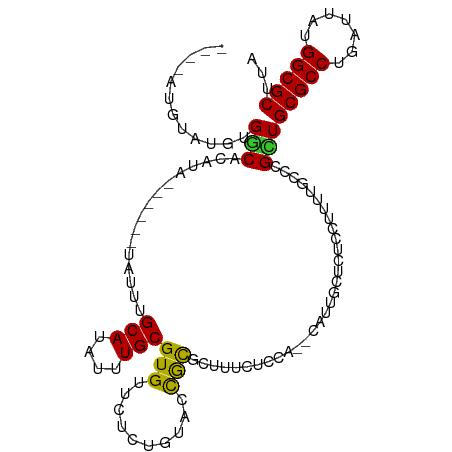
>dm3.chr2L 17100674 96 + 23011544 ---UUUCCCAUUUCACUGCCCU--CCUGCUUUUCCUCCACUUA--UUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCG-GGCAAAAGGAGAGCA ---..............((.((--(((..((.(((..((((((--......))))))..........(((((.......)))))...)-)).)).))))).)). ( -28.70, z-score = -3.28, R) >droSim1.chr2L 16785385 96 + 22036055 ---UUUCCCAUUUCACUGCCCU--CCUAGUUUUCCUCCACUUA--UUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCG-GGCAAAAGGAGAGCA ---..............((.((--(((..((.(((..((((((--......))))))..........(((((.......)))))...)-)).)).))))).)). ( -27.70, z-score = -3.50, R) >droSec1.super_7 732190 96 + 3727775 ---UUUCCCAUUUCACUGCCCU--CCUGGUUUUCCUCCACUUA--UUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCG-GGCAAAAGGAGAGCA ---..............((.((--(((..((.(((..((((((--......))))))..........(((((.......)))))...)-)).)).))))).)). ( -28.90, z-score = -3.07, R) >droYak2.chr2R 17140473 96 - 21139217 ---UUUCCCAUUUCACUGCCCU--CCCGGUUUUCCUGCACUUA--UUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCG-GGUAAGCGGGGAGGA ---................(((--(((.((((.((((((((((--......)))))...........(((((.......))))).)))-)).)))).)))))). ( -40.30, z-score = -5.22, R) >droEre2.scaffold_4845 15452936 96 - 22589142 ---UUUCCCAUUUCACUGCCCU--CCCGGUCUUCCUCCACUUA--UUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCG-GGUAAAAGGAGAGCA ---..............((.((--((...(..(((..((((((--......))))))..........(((((.......)))))...)-))..)..)))).)). ( -26.40, z-score = -2.55, R) >droAna3.scaffold_12916 12431427 101 - 16180835 ---UUUCCUGUGUUCCUGCCCCAGCCCUUCUUUCCAUCGCCUCCUUUUUUUUAAGUGUUUUCCUUAAGCGCCGUAAUCAGGCGCAGCGCGGCAGACAGAGAGCG ---.(((((((....(((((...((.........................(((((.(....))))))(((((.......))))).))..))))))))).))).. ( -26.30, z-score = -1.73, R) >dp4.chr4_group3 275239 92 - 11692001 ---UAUAUCGACUGAUAGCCCUCUUCCUUGUUUCUUUC--------CGUUUUAAGUGUUUUCCUUAAGCGCCAUAAUCAGGCGCAACG-GGAAAAACGAAAGAA ---..((((....))))....((((..((((((.((((--------(((((((((.(....))))))(((((.......)))))))))-)))))))))))))). ( -25.60, z-score = -2.90, R) >droPer1.super_8 1315922 92 - 3966273 ---UAUAUCGACUGAUAGCCCUCUUCCUUGUUUCUUUC--------CGUUUUAAGUGUUUUCCUUAAGCGCCAUAAUCAGGCGCAACG-AGAAAAACGAAAGAA ---..((((....))))....((((..((((((.((..--------(((((((((.(....))))))(((((.......)))))))))-..)))))))))))). ( -20.50, z-score = -1.87, R) >droWil1.scaffold_180703 2905107 82 - 3946847 ----------------CCUUGUUGUUGCUGUUGUUGUUAGCUUC-CUUUUUUAAGUGUUU-CUUUAAGCGCCAUAAUCAGGCGCAGCG----AGAGCGAAAAUA ----------------...((((.(((((.(((((....((((.-.......))))....-......(((((.......)))))))))----).))))).)))) ( -22.00, z-score = -1.64, R) >droGri2.scaffold_15252 16305509 98 + 17193109 UUCUGUUUCUCUUCACUUUUUUUCUUGCAGUUGUAUCUGGUUUG-GCUUUAUAAGUGUUU-CUUUAAGCGCCAUAAUCAAGCGCACCA----AGAACGCAAAUA .....................((((((....((..(.(((((((-((((((.(((.....-))))))).)))).))))).)..)).))----))))........ ( -15.20, z-score = 0.28, R) >consensus ___UUUCCCAUUUCACUGCCCU__CCCGGUUUUCCUCCACUUA__UUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCG_GGCAAAAGGAGAGCA ..............................((((................(((((.......)))))(((((.......))))).............))))... ( -9.56 = -9.12 + -0.44)
| Location | 17,100,674 – 17,100,770 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 69.23 |
| Shannon entropy | 0.65789 |
| G+C content | 0.44027 |
| Mean single sequence MFE | -27.40 |
| Consensus MFE | -10.01 |
| Energy contribution | -10.19 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.57 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956115 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17100674 96 - 23011544 UGCUCUCCUUUUGCC-CGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAA--UAAGUGGAGGAAAAGCAGG--AGGGCAGUGAAAUGGGAAA--- ((((((((((((..(-(...(((((.......)))))..........((((((......--))))))..))..))).)))--)))))).............--- ( -33.20, z-score = -3.03, R) >droSim1.chr2L 16785385 96 - 22036055 UGCUCUCCUUUUGCC-CGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAA--UAAGUGGAGGAAAACUAGG--AGGGCAGUGAAAUGGGAAA--- (((((((((.((..(-(...(((((.......)))))..........((((((......--))))))..))..))..)))--)))))).............--- ( -32.30, z-score = -3.33, R) >droSec1.super_7 732190 96 - 3727775 UGCUCUCCUUUUGCC-CGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAA--UAAGUGGAGGAAAACCAGG--AGGGCAGUGAAAUGGGAAA--- ((((((((((((..(-(...(((((.......)))))..........((((((......--))))))..)).)))..)))--)))))).............--- ( -31.60, z-score = -2.75, R) >droYak2.chr2R 17140473 96 + 21139217 UCCUCCCCGCUUACC-CGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAA--UAAGUGCAGGAAAACCGGG--AGGGCAGUGAAAUGGGAAA--- (((((((.(.((.((-.((.(((((.......)))))...........(((((......--))))))).)).)).).)))--))))...............--- ( -28.80, z-score = -1.54, R) >droEre2.scaffold_4845 15452936 96 + 22589142 UGCUCUCCUUUUACC-CGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAA--UAAGUGGAGGAAGACCGGG--AGGGCAGUGAAAUGGGAAA--- ((((((((((((.((-(...(((((.......)))))..........((((((......--))))))).)).)))..)))--)))))).............--- ( -31.30, z-score = -2.67, R) >droAna3.scaffold_12916 12431427 101 + 16180835 CGCUCUCUGUCUGCCGCGCUGCGCCUGAUUACGGCGCUUAAGGAAAACACUUAAAAAAAAGGAGGCGAUGGAAAGAAGGGCUGGGGCAGGAACACAGGAAA--- .(((((..((((.((((((((((((.......)))))(((((.......))))).........)))).)))......)))).)))))..............--- ( -30.40, z-score = -0.68, R) >dp4.chr4_group3 275239 92 + 11692001 UUCUUUCGUUUUUCC-CGUUGCGCCUGAUUAUGGCGCUUAAGGAAAACACUUAAAACG--------GAAAGAAACAAGGAAGAGGGCUAUCAGUCGAUAUA--- (((((((.(((((.(-(((((((((.......)))))(((((.......)))))))))--------).)))))....)))))))(((.....)))......--- ( -28.30, z-score = -2.82, R) >droPer1.super_8 1315922 92 + 3966273 UUCUUUCGUUUUUCU-CGUUGCGCCUGAUUAUGGCGCUUAAGGAAAACACUUAAAACG--------GAAAGAAACAAGGAAGAGGGCUAUCAGUCGAUAUA--- (((((((.((((((.-(((((((((.......)))))(((((.......)))))))))--------)))))).....)))))))(((.....)))......--- ( -27.30, z-score = -2.75, R) >droWil1.scaffold_180703 2905107 82 + 3946847 UAUUUUCGCUCU----CGCUGCGCCUGAUUAUGGCGCUUAAAG-AAACACUUAAAAAAG-GAAGCUAACAACAACAGCAACAACAAGG---------------- .......(((..----(...(((((.......)))))...(((-.....)))......)-..))).......................---------------- ( -14.20, z-score = -0.45, R) >droGri2.scaffold_15252 16305509 98 - 17193109 UAUUUGCGUUCU----UGGUGCGCUUGAUUAUGGCGCUUAAAG-AAACACUUAUAAAGC-CAAACCAGAUACAACUGCAAGAAAAAAAGUGAAGAGAAACAGAA ..((((..((((----(....(((((.....((((..((((((-.....))).))).))-))...(((......))).........)))))..))))).)))). ( -16.60, z-score = -0.00, R) >consensus UGCUCUCCUUUUGCC_CGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAA__UAAGUGGAGGAAAAACGGG__AGGGCAGUGAAAUGGGAAA___ .....((((((.........(((((.......)))))...(((......)))..............))))))................................ (-10.01 = -10.19 + 0.18)
| Location | 17,100,702 – 17,100,808 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 66.53 |
| Shannon entropy | 0.67228 |
| G+C content | 0.43307 |
| Mean single sequence MFE | -26.26 |
| Consensus MFE | -12.24 |
| Energy contribution | -11.58 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
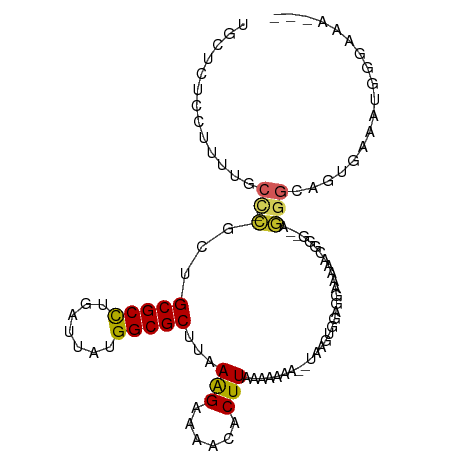
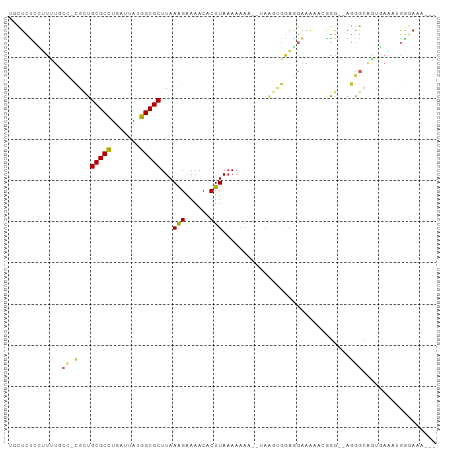
>dm3.chr2L 17100702 106 + 23011544 --------CCUCCACUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGCAAAA--GGAGAGCAAUGUGGAGAAUGCGCGGUACAGAGAACACGCAAAUAUG --------.(((((((((......))).((((((((((..(((((.......))))).(....)..))--))))))))..))))))..((((.(..........).))))...... ( -31.40, z-score = -2.15, R) >droSim1.chr2L 16785413 106 + 22036055 --------CCUCCACUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGCAAAA--GGAGAGCAAUGUGGAGAAAGCGCGGUACAGAGAACACGCAAAUAUG --------.(((((((((......))).((((((((((..(((((.......))))).(....)..))--))))))))..)))))).....(((((.......)).)))....... ( -30.70, z-score = -2.08, R) >droSec1.super_7 732218 106 + 3727775 --------CCUCCACUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGCAAAA--GGAGAGCAAUGUGGAGAAAGCGCGGUACAGAGAACACGCAAAUAUG --------.(((((((((......))).((((((((((..(((((.......))))).(....)..))--))))))))..)))))).....(((((.......)).)))....... ( -30.70, z-score = -2.08, R) >droYak2.chr2R 17140501 106 - 21139217 --------CCUGCACUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGUAAGC--GGGGAGGAAUGUGGAGAAAGCGCGGUACAGAGAACACGCAAAUAUG --------.(((((((((......)))))((((((((((.(((((.......))))).((......))--...........))))))))))))))..................... ( -29.80, z-score = -1.26, R) >droEre2.scaffold_4845 15452964 106 - 22589142 --------CCUCCACUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGUAAAA--GGAGAGCAAUGUGGAGAAAGCGCGGCACGGAGAACACGCAAAUAUG --------.(((((((((......)))))((((((((((.(((((.......))))).((........--.....))....)))))))))).......)))).............. ( -29.82, z-score = -1.47, R) >droAna3.scaffold_12916 12431457 109 - 16180835 ------CCAUCGCCUCCUUUUUUUUAAGUGUUUUCCUUAAGCGCCGUAAUCAGGCGCAGCGCGGCAGAC-AGAGAGCGGUUGAAUGGAAGAGCAGGGCAGAAGGCACGCAAAUAUG ------((.(((((((((((..(((((.(((((((.....(((((.......))))).((...))....-.))))))).)))))..)))).)..)))).)).))............ ( -32.20, z-score = -0.20, R) >dp4.chr4_group3 275267 104 - 11692001 ------------UUCUUUCCGUUUUAAGUGUUUUCCUUAAGCGCCAUAAUCAGGCGCAACGGGAAAAACGAAAGAAAGAGAUAGAGAAAUAGCGGCAUAGAAAACACGCAAAUAUG ------------...((((..((((.....(((((((...(((((.......)))))...))))))).(....)...))))..))))....(((............)))....... ( -25.30, z-score = -2.38, R) >droPer1.super_8 1315950 104 - 3966273 ------------UUCUUUCCGUUUUAAGUGUUUUCCUUAAGCGCCAUAAUCAGGCGCAACGAGAAAAACGAAAGAAAGAGAUAGAGAAAUAGCGGCAUAGAAAACACGCAAAUAUG ------------((((.((..((((...((((((.(((..(((((.......)))))...))).))))))...))))..)).)))).....(((............)))....... ( -23.00, z-score = -2.09, R) >droWil1.scaffold_180703 2905125 86 - 3946847 --------UGUUAGCUUCCUUUUUUAAGUGUUU-CUUUAAGCGCCAUAAUCAGGCGCAGCGAGAGCGA---------------------AAAUAUUUUAAGCAACACGCAAAUAUG --------((((.((((........((((((((-(.....(((((.......))))).((....))).---------------------)))))))).)))))))).......... ( -20.30, z-score = -1.47, R) >droVir3.scaffold_13246 1767046 93 - 2672878 GUUGUUGUAUCUGGUU-GGCUUUUUAAGUGUUU-CUUUAAGCGCCAUAAUCAGGCGCACCAAGAGCGC---------------------AAAUAUUUUAAGCGACACGCAAAUAUG ((((((((..((((((-(((.(((.(((.....-))).))).))))..)))))((((.......))))---------------------...........)))))).))....... ( -23.90, z-score = -1.16, R) >droMoj3.scaffold_6500 5015875 84 + 32352404 ----------CUGGUUGGCUUUUUUAAGUGUUU-CUUUAAGCGCCAUAAUCAGGCGCACCAAGAGCGC---------------------AAAUAUUUUAAGCGACACGCAAAUAUG ----------...((((.(((...((..(((((-(((...(((((.......)))))...))))).))---------------------)..))....)))))))........... ( -19.00, z-score = -0.47, R) >droGri2.scaffold_15252 16305535 94 + 17193109 GCAGUUGUAUCUGGUUUGGCUUUAUAAGUGUUU-CUUUAAGCGCCAUAAUCAAGCGCACCAAGAACGC---------------------AAAUAUUUUAAGCGACACGCAAAUAUG ......((((.((((.(.((((.(((..(((((-(((...((((.........))))...))))).))---------------------)..)))...)))).).)).)).)))). ( -19.00, z-score = 0.00, R) >consensus ________CCUCCACUUAUUUUUUUAAGUGUUUUCUUUAAGCGCCAUAAUCAGGCGCAGCGGGAAAAA__GGAGAGCAAUGUGGAGAAAGAGCGGUACAGAGAACACGCAAAUAUG ...........................(((((........(((((.......)))))...................................(......)..)))))......... (-12.24 = -11.58 + -0.66)

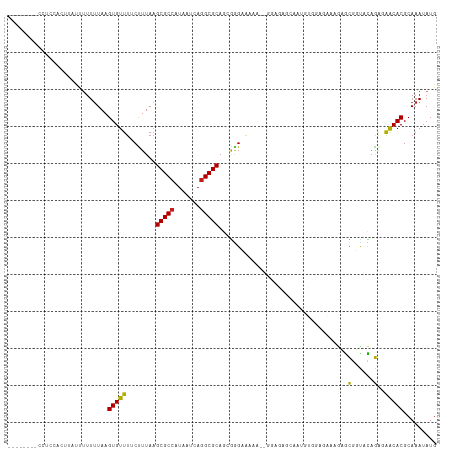
| Location | 17,100,702 – 17,100,808 |
|---|---|
| Length | 106 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 66.53 |
| Shannon entropy | 0.67228 |
| G+C content | 0.43307 |
| Mean single sequence MFE | -23.45 |
| Consensus MFE | -9.91 |
| Energy contribution | -9.81 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855218 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
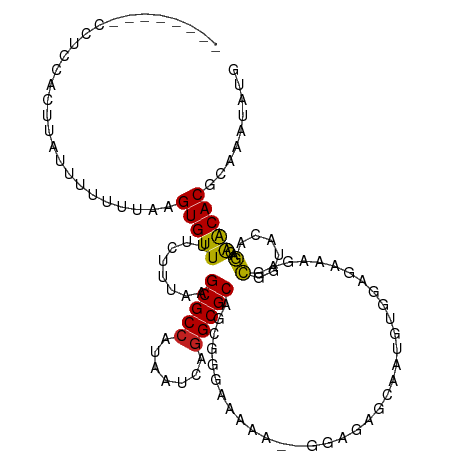
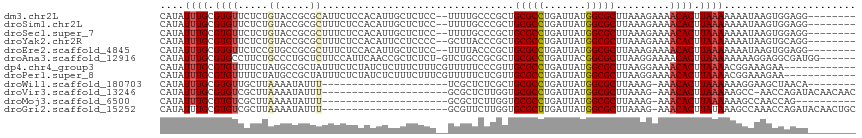
>dm3.chr2L 17100702 106 - 23011544 CAUAUUUGCGUGUUCUCUGUACCGCGCAUUCUCCACAUUGCUCUCC--UUUUGCCCGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAAUAAGUGGAGG-------- ......((((((..........))))))..((((((...((.....--....)).....(((((.......))))).........................)))))).-------- ( -27.00, z-score = -1.91, R) >droSim1.chr2L 16785413 106 - 22036055 CAUAUUUGCGUGUUCUCUGUACCGCGCUUUCUCCACAUUGCUCUCC--UUUUGCCCGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAAUAAGUGGAGG-------- .......(((((..........)))))...((((((...((.....--....)).....(((((.......))))).........................)))))).-------- ( -25.80, z-score = -1.60, R) >droSec1.super_7 732218 106 - 3727775 CAUAUUUGCGUGUUCUCUGUACCGCGCUUUCUCCACAUUGCUCUCC--UUUUGCCCGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAAUAAGUGGAGG-------- .......(((((..........)))))...((((((...((.....--....)).....(((((.......))))).........................)))))).-------- ( -25.80, z-score = -1.60, R) >droYak2.chr2R 17140501 106 + 21139217 CAUAUUUGCGUGUUCUCUGUACCGCGCUUUCUCCACAUUCCUCCCC--GCUUACCCGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAAUAAGUGCAGG-------- .......(((((..........)))))(((((..............--((......)).(((((.......)))))....))))).((((((......))))))....-------- ( -22.50, z-score = -1.43, R) >droEre2.scaffold_4845 15452964 106 + 22589142 CAUAUUUGCGUGUUCUCCGUGCCGCGCUUUCUCCACAUUGCUCUCC--UUUUACCCGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAAUAAGUGGAGG-------- .......(((((..........)))))...((((((...((.....--........)).(((((.......))))).........................)))))).-------- ( -25.22, z-score = -1.31, R) >droAna3.scaffold_12916 12431457 109 + 16180835 CAUAUUUGCGUGCCUUCUGCCCUGCUCUUCCAUUCAACCGCUCUCU-GUCUGCCGCGCUGCGCCUGAUUACGGCGCUUAAGGAAAACACUUAAAAAAAAGGAGGCGAUGG------ ........((((((((((((...))..((((.......(((.....-.......)))..(((((.......)))))....))))..............))))))).))).------ ( -25.80, z-score = 0.56, R) >dp4.chr4_group3 275267 104 + 11692001 CAUAUUUGCGUGUUUUCUAUGCCGCUAUUUCUCUAUCUCUUUCUUUCGUUUUUCCCGUUGCGCCUGAUUAUGGCGCUUAAGGAAAACACUUAAAACGGAAAGAA------------ .......(((.((.......)))))............(((((((...((((((((....(((((.......)))))....)))))).)).......))))))).------------ ( -24.60, z-score = -2.39, R) >droPer1.super_8 1315950 104 + 3966273 CAUAUUUGCGUGUUUUCUAUGCCGCUAUUUCUCUAUCUCUUUCUUUCGUUUUUCUCGUUGCGCCUGAUUAUGGCGCUUAAGGAAAACACUUAAAACGGAAAGAA------------ .......(((.((.......)))))...............(((((((((((((((....(((((.......)))))...)))))))).(.......))))))))------------ ( -25.30, z-score = -2.74, R) >droWil1.scaffold_180703 2905125 86 + 3946847 CAUAUUUGCGUGUUGCUUAAAAUAUUU---------------------UCGCUCUCGCUGCGCCUGAUUAUGGCGCUUAAAG-AAACACUUAAAAAAGGAAGCUAACA-------- ....((((.(((((.(((.........---------------------..((....)).(((((.......)))))...)))-.))))).))))..............-------- ( -18.00, z-score = -0.52, R) >droVir3.scaffold_13246 1767046 93 + 2672878 CAUAUUUGCGUGUCGCUUAAAAUAUUU---------------------GCGCUCUUGGUGCGCCUGAUUAUGGCGCUUAAAG-AAACACUUAAAAAGCC-AACCAGAUACAACAAC .........(((((((...........---------------------))....((((((((((.......)))))...(((-.....))).....)))-))...)))))...... ( -20.70, z-score = -1.04, R) >droMoj3.scaffold_6500 5015875 84 - 32352404 CAUAUUUGCGUGUCGCUUAAAAUAUUU---------------------GCGCUCUUGGUGCGCCUGAUUAUGGCGCUUAAAG-AAACACUUAAAAAAGCCAACCAG---------- ....((((.(((((((...........---------------------))).((((...(((((.......)))))...)))-).)))).))))............---------- ( -19.30, z-score = -0.88, R) >droGri2.scaffold_15252 16305535 94 - 17193109 CAUAUUUGCGUGUCGCUUAAAAUAUUU---------------------GCGUUCUUGGUGCGCUUGAUUAUGGCGCUUAAAG-AAACACUUAUAAAGCCAAACCAGAUACAACUGC ..((((((.((...((((...(((..(---------------------(..(((((...(((((.......)))))...)))-)).))..))).))))...))))))))....... ( -21.40, z-score = -0.85, R) >consensus CAUAUUUGCGUGUUCUCUAUACCGCUCUUUCUCCACAUUGCUCUCC__UCUUUCCCGCUGCGCCUGAUUAUGGCGCUUAAAGAAAACACUUAAAAAAAUAAGUGAAGG________ ....((((.((((.....((.....))................................(((((.......))))).........)))).))))...................... ( -9.91 = -9.81 + -0.10)
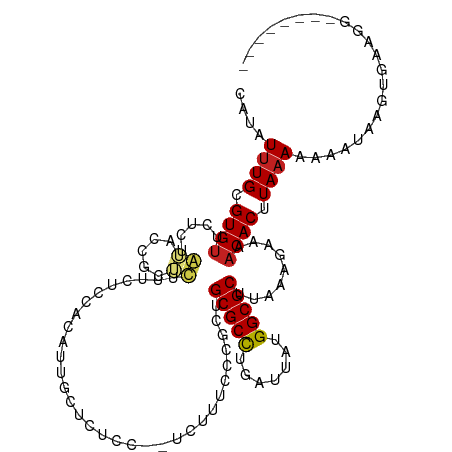
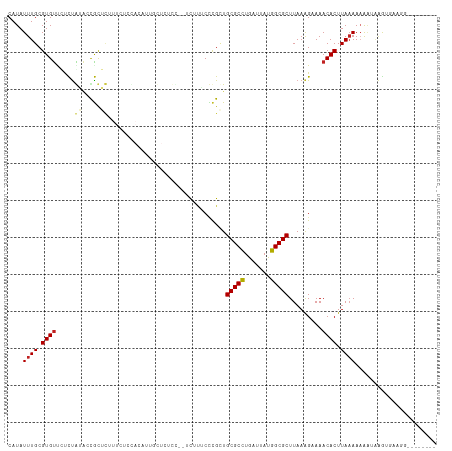
| Location | 17,100,731 – 17,100,830 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.46 |
| Shannon entropy | 0.51898 |
| G+C content | 0.46429 |
| Mean single sequence MFE | -24.71 |
| Consensus MFE | -12.28 |
| Energy contribution | -12.28 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793720 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
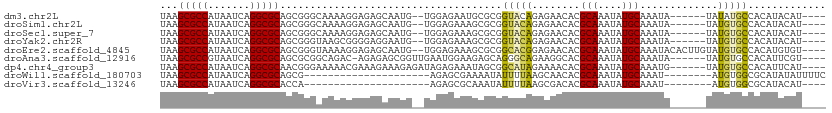
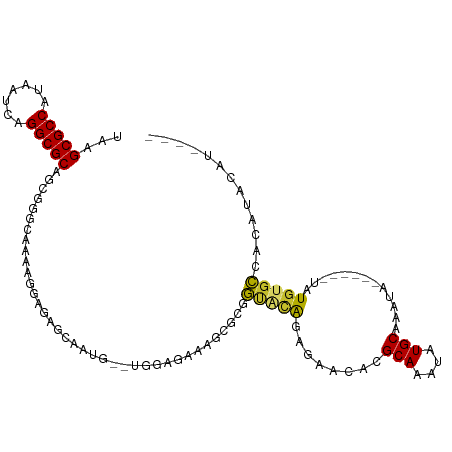
>dm3.chr2L 17100731 99 + 23011544 UAAGCGCCAUAAUCAGGCGCAGCGGGCAAAAGGAGAGCAAUG--UGGAGAAUGCGCGGUACAGAGAACACGCAAAUAUGCAAAUA------UAUAUGCCACAUACAU---- ...(((((.......))))).((...(.....)...)).(((--(((....((((.(..........).)))).((((....)))------).....))))))....---- ( -23.90, z-score = -1.10, R) >droSim1.chr2L 16785442 99 + 22036055 UAAGCGCCAUAAUCAGGCGCAGCGGGCAAAAGGAGAGCAAUG--UGGAGAAAGCGCGGUACAGAGAACACGCAAAUAUGCAAAUA------UAUGUGCCACAUACAU---- ...(((((.......))))).((...(.....)...)).(((--(((.....(((((((.......)).)))..((((....)))------)..)).))))))....---- ( -23.60, z-score = -0.91, R) >droSec1.super_7 732247 99 + 3727775 UAAGCGCCAUAAUCAGGCGCAGCGGGCAAAAGGAGAGCAAUG--UGGAGAAAGCGCGGUACAGAGAACACGCAAAUAUGCAAAUA------UAUGUGCCACAUACAU---- ...(((((.......))))).((...(.....)...)).(((--(((.....(((((((.......)).)))..((((....)))------)..)).))))))....---- ( -23.60, z-score = -0.91, R) >droYak2.chr2R 17140530 99 - 21139217 UAAGCGCCAUAAUCAGGCGCAGCGGGUAAGCGGGGAGGAAUG--UGGAGAAAGCGCGGUACAGAGAACACGCAAAUAUGCAAAUA------UAUGUGCCACAUACAU---- ...(((((.......))))).((......))......(.(((--(((.....(((((((.......)).)))..((((....)))------)..)).)))))).)..---- ( -24.50, z-score = -0.87, R) >droEre2.scaffold_4845 15452993 105 - 22589142 UAAGCGCCAUAAUCAGGCGCAGCGGGUAAAAGGAGAGCAAUG--UGGAGAAAGCGCGGCACGGAGAACACGCAAAUAUGCAAAUACACUUGUAUGUGCCACAUGUGU---- ...(((((.......)))))................((((((--(((.....(((.(..........).)))..((((((((......)))))))).)))))).)))---- ( -28.60, z-score = -0.53, R) >droAna3.scaffold_12916 12431488 100 - 16180835 UAAGCGCCGUAAUCAGGCGCAGCGCGGCAGAC-AGAGAGCGGUUGAAUGGAAGAGCAGGGCAGAAGGCACGCAAAUAUGCAAAUA------UAUGUGCCACAUUCGU---- ...(((((.......)))))..(((..(....-.)...)))...(((((.....((...))....((((((...((((....)))------).)))))).)))))..---- ( -28.30, z-score = -0.62, R) >dp4.chr4_group3 275292 101 - 11692001 UAAGCGCCAUAAUCAGGCGCAACGGGAAAAACGAAAGAAAGAGAUAGAGAAAUAGCGGCAUAGAAAACACGCAAAUAUGCAAAUG------UAUGUGCCACAUUCAU---- ...(((((.......))))).....(((...(....).................(.((((((....(((.(((....)))...))------).)))))).).)))..---- ( -23.20, z-score = -3.05, R) >droWil1.scaffold_180703 2905153 82 - 3946847 UAAGCGCCAUAAUCAGGCGCAGCG---------------------AGAGCGAAAAUAUUUUAAGCAACACGCAAAUAUGCAAAU--------AUGUGGCGCAUAUAUUUUC ...(((((.......))))).((.---------------------...))((((((((.....((.(((.(((....)))....--------.))).))....)))))))) ( -23.10, z-score = -2.61, R) >droVir3.scaffold_13246 1767081 78 - 2672878 UAAGCGCCAUAAUCAGGCGCACCA---------------------AGAGCGCAAAUAUUUUAAGCGACACGCAAAUAUGCAAAU--------AUGUGGCGCAUACAU---- ...((((((((.....((((....---------------------...))))..((((((...(((...)))))))))......--------.))))))))......---- ( -23.60, z-score = -3.02, R) >consensus UAAGCGCCAUAAUCAGGCGCAGCGGGCAAAAGGAGAGCAAUG__UGGAGAAAGCGCGGUACAGAGAACACGCAAAUAUGCAAAUA______UAUGUGCCACAUACAU____ ...(((((.......)))))..................................................(((....)))............................... (-12.28 = -12.28 + 0.00)
| Location | 17,100,731 – 17,100,830 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.46 |
| Shannon entropy | 0.51898 |
| G+C content | 0.46429 |
| Mean single sequence MFE | -23.12 |
| Consensus MFE | -14.62 |
| Energy contribution | -13.92 |
| Covariance contribution | -0.70 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.08 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830230 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
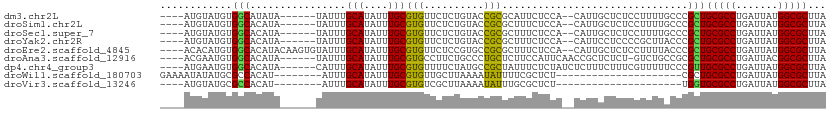
>dm3.chr2L 17100731 99 - 23011544 ----AUGUAUGUGGCAUAUA------UAUUUGCAUAUUUGCGUGUUCUCUGUACCGCGCAUUCUCCA--CAUUGCUCUCCUUUUGCCCGCUGCGCCUGAUUAUGGCGCUUA ----..((((((((((....------....))).....((((((..........)))))).....))--)).)))................(((((.......)))))... ( -24.50, z-score = -1.15, R) >droSim1.chr2L 16785442 99 - 22036055 ----AUGUAUGUGGCACAUA------UAUUUGCAUAUUUGCGUGUUCUCUGUACCGCGCUUUCUCCA--CAUUGCUCUCCUUUUGCCCGCUGCGCCUGAUUAUGGCGCUUA ----..((((((((((....------....)))......(((((..........)))))......))--)).)))................(((((.......)))))... ( -22.40, z-score = -0.69, R) >droSec1.super_7 732247 99 - 3727775 ----AUGUAUGUGGCACAUA------UAUUUGCAUAUUUGCGUGUUCUCUGUACCGCGCUUUCUCCA--CAUUGCUCUCCUUUUGCCCGCUGCGCCUGAUUAUGGCGCUUA ----..((((((((((....------....)))......(((((..........)))))......))--)).)))................(((((.......)))))... ( -22.40, z-score = -0.69, R) >droYak2.chr2R 17140530 99 + 21139217 ----AUGUAUGUGGCACAUA------UAUUUGCAUAUUUGCGUGUUCUCUGUACCGCGCUUUCUCCA--CAUUCCUCCCCGCUUACCCGCUGCGCCUGAUUAUGGCGCUUA ----....((((((((....------....)))......(((((..........)))))......))--)))........((......)).(((((.......)))))... ( -22.20, z-score = -1.07, R) >droEre2.scaffold_4845 15452993 105 + 22589142 ----ACACAUGUGGCACAUACAAGUGUAUUUGCAUAUUUGCGUGUUCUCCGUGCCGCGCUUUCUCCA--CAUUGCUCUCCUUUUACCCGCUGCGCCUGAUUAUGGCGCUUA ----......(((((((((((....))))..(((....))).........)))))))((........--....))................(((((.......)))))... ( -27.40, z-score = -1.42, R) >droAna3.scaffold_12916 12431488 100 + 16180835 ----ACGAAUGUGGCACAUA------UAUUUGCAUAUUUGCGUGCCUUCUGCCCUGCUCUUCCAUUCAACCGCUCUCU-GUCUGCCGCGCUGCGCCUGAUUACGGCGCUUA ----..(((((.(((((...------.....(((....))))))))....((...)).....)))))...(((.....-.......)))..(((((.......)))))... ( -22.60, z-score = 0.04, R) >dp4.chr4_group3 275292 101 + 11692001 ----AUGAAUGUGGCACAUA------CAUUUGCAUAUUUGCGUGUUUUCUAUGCCGCUAUUUCUCUAUCUCUUUCUUUCGUUUUUCCCGUUGCGCCUGAUUAUGGCGCUUA ----..(((.((((((...(------(((..(((....)))))))......))))))...)))............................(((((.......)))))... ( -23.80, z-score = -2.96, R) >droWil1.scaffold_180703 2905153 82 + 3946847 GAAAAUAUAUGCGCCACAU--------AUUUGCAUAUUUGCGUGUUGCUUAAAAUAUUUUCGCUCU---------------------CGCUGCGCCUGAUUAUGGCGCUUA ((((((((.(..((.((((--------....(((....))))))).))...).))))))))((...---------------------.)).(((((.......)))))... ( -22.10, z-score = -1.67, R) >droVir3.scaffold_13246 1767081 78 + 2672878 ----AUGUAUGCGCCACAU--------AUUUGCAUAUUUGCGUGUCGCUUAAAAUAUUUGCGCUCU---------------------UGGUGCGCCUGAUUAUGGCGCUUA ----......((((((...--------(((.(((((((((((...)))...))))))..((((...---------------------..))))))..)))..))))))... ( -20.70, z-score = -0.13, R) >consensus ____AUGUAUGUGGCACAUA______UAUUUGCAUAUUUGCGUGUUCUCUGUACCGCGCUUUCUCCA__CAUUGCUCUCCUUUUGCCCGCUGCGCCUGAUUAUGGCGCUUA ............(((................(((....)))(((..........)))...............................)))(((((.......)))))... (-14.62 = -13.92 + -0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:47:15 2011