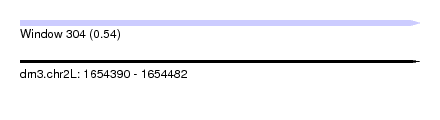
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,654,390 – 1,654,482 |
| Length | 92 |
| Max. P | 0.538470 |

| Location | 1,654,390 – 1,654,482 |
|---|---|
| Length | 92 |
| Sequences | 8 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 73.79 |
| Shannon entropy | 0.50282 |
| G+C content | 0.48944 |
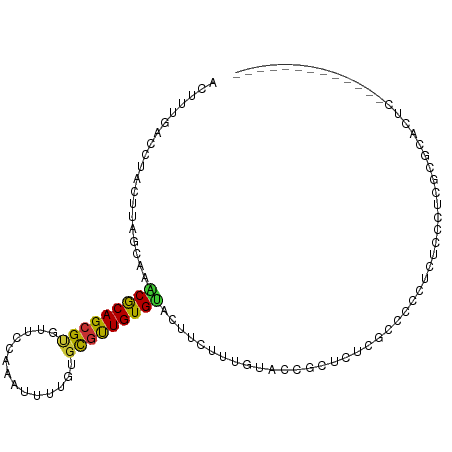
| Mean single sequence MFE | -15.62 |
| Consensus MFE | -10.19 |
| Energy contribution | -9.89 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.67 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.538470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
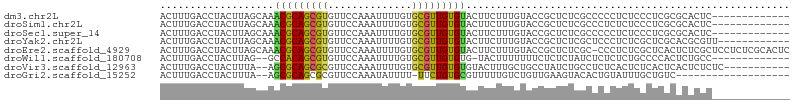
>dm3.chr2L 1654390 92 + 23011544 ACUUUGACCUACUUAGCAAACGCAGCGUGUUCCAAAUUUUGUGCGUUGUGUACUUCUUUGUACCGCUCUCGCCCCCUCUCCCUCGCGCACUC------------- ...............((....)).(((((...........(.(((..((((((......))).)))...))))..........)))))....------------- ( -16.50, z-score = -1.27, R) >droSim1.chr2L 1688296 92 + 22036055 ACUUUGACCUACUUAGCAAACGCAGCGUGUUCCAAAUUUUGUGCGUUGUGUACUUCUUUGUACCGCUCUCGCCCUCUCUCCCUCGCGCACUC------------- ...............((....)).(((((...........(.(((..((((((......))).)))...))).).........)))))....------------- ( -16.75, z-score = -1.22, R) >droSec1.super_14 1596948 92 + 2068291 ACUUUGACCUACUUAGCAAACGCAGCGUGUUCCAAAUUUUGUGCGUUGUGUACUUCUUUGUACCGCUCUCGCCCCCUCUCCCUCGCGCACUC------------- ...............((....)).(((((...........(.(((..((((((......))).)))...))))..........)))))....------------- ( -16.50, z-score = -1.27, R) >droYak2.chr2L 1629839 93 + 22324452 ACUUUGACCUACUUAGCAAACGCAGCGUGUUCCAAAUUUUGUGCGUUGUGUACUUCUUUGUACCGCUCUCGCUCCCUCUCGCUCGCACGCGUU------------ ...............((....)).((((((............(((..((((((......))).)))...)))............))))))...------------ ( -19.55, z-score = -0.72, R) >droEre2.scaffold_4929 1704939 104 + 26641161 ACUUUGACCUACUUAGCAAACGCAGCGUGUUCCAAAUUUUGUGCGUUGUGUACUUCUUUGUACCGCUCUCGC-CCCUCUCGCUCACUCUCGCUCCUCUCGCACUC ..............(((....((((((..(..........)..))))))((((......)))).)))...((-.......((........)).......)).... ( -15.94, z-score = -0.52, R) >droWil1.scaffold_180708 3519518 89 + 12563649 ACUUUGACCUACUUAG--GCCACAGCGUGUUCCAAAUUUUGUGCGUUGUGUG-UACUUUUUUUCUCUCUAUCUCUCUCUGCCCCACUCUGCC------------- ................--(((((((((..(..........)..))))))).)-)......................................------------- ( -12.30, z-score = -0.74, R) >droVir3.scaffold_12963 7746940 92 + 20206255 ACUUUGACCUACUUUA--AGCGCAGCGCGUUCCAAAUUUUGUGCGUUGUGUGUACUUUGCUGCCUAUCUGCCUCUCACUCUCACUCACUCUCUC----------- ................--.((((((((((............))))))))))...........................................----------- ( -13.00, z-score = 0.02, R) >droGri2.scaffold_15252 10830614 83 + 17193109 ACUUUGACCUACUUUA--AGCGCAGCGCGUUCCAAAUAUUUU-UUCUGUGCGUUUUUGUCUGUUGAAGUACACUGUAUUUGCUGUC------------------- ((((..((..((....--.((((((.................-..))))))......))..))..)))).................------------------- ( -14.41, z-score = -0.03, R) >consensus ACUUUGACCUACUUAGCAAACGCAGCGUGUUCCAAAUUUUGUGCGUUGUGUACUUCUUUGUACCGCUCUCGCCCCCUCUCCCUCGCGCACUC_____________ ...................(((((((((..............)))))))))...................................................... (-10.19 = -9.89 + -0.30)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:53 2011