
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,086,798 – 17,086,895 |
| Length | 97 |
| Max. P | 0.794660 |

| Location | 17,086,798 – 17,086,895 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 82.90 |
| Shannon entropy | 0.34070 |
| G+C content | 0.41635 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -21.72 |
| Energy contribution | -21.08 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.794660 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 17086798 97 + 23011544 ------------GCCUGGGGCGUGGCACAUGCAAAUGACACGCCUGAUGCUGCGUAAAUAAUUUUAGCGCU---CUCUC----UUUUGGCAUAAAAUGCUAAAUAAACAUUUUAUU ------------((.(.(((((((...(((....))).))))))).).)).((((.((.....)).)))).---.....----.((((((((...))))))))............. ( -26.60, z-score = -1.76, R) >droSim1.chr2L 16770973 97 + 22036055 ------------GGCUGGGGCGUGGCACAUGCAAAUGACACGCCUGAUGCUGCGUAAAUAAUUUUAGCGCU---CUCUC----UUUUGGCAUAAAAUGCUAAAUAAACAUUUUAUU ------------((((.(((((((...(((....))).))))))).).)))((((.((.....)).)))).---.....----.((((((((...))))))))............. ( -27.10, z-score = -1.92, R) >droSec1.super_7 718537 97 + 3727775 ------------UGCUGGGGCGUGGCACAUGCAAAUGACACGCCUGAUGCUGCGUAAAUAAUUUUAGCGCU---CUCUC----UUUUGGCAUAAAAUGCUAAAUAAACAUUUUAUU ------------.(((.(((((((...(((....))).))))))).).)).((((.((.....)).)))).---.....----.((((((((...))))))))............. ( -26.00, z-score = -1.60, R) >droYak2.chr2R 17126704 101 - 21139217 ------------GCUGGGGGCGUGGCACAUGCAAAUGACACGCCUGAUGCUGCGUAAAUAAUUUUAGCGCU---CUCUCUCUCUUUUGGCAUAAAAUGCUAAAUAAACAUUUUAUU ------------((...(((((((...(((....))).)))))))...)).((((.((.....)).)))).---..........((((((((...))))))))............. ( -25.30, z-score = -1.35, R) >droEre2.scaffold_4845 15439093 97 - 22589142 ------------GCUGGGGGCGUGGCACAUGCAAAUGACACGCCUGAUGCUGCGUAAAUAAUUUUAGCGCU---CUCUC----UUUUGGCAUAAAAUGCUAAAUAAACAUUUUAUU ------------((...(((((((...(((....))).)))))))...)).((((.((.....)).)))).---.....----.((((((((...))))))))............. ( -25.30, z-score = -1.28, R) >droAna3.scaffold_12916 12418351 95 - 16180835 ------------UCCGAAGGCGUGGCACAUGCAAAUGACACGCCUGAUGCUGCGUAAAUAAUUUUAGCGCU---CUCUC------UUGGCAUAAAAUGCAAAAUAAACAUUUUAUU ------------.(((((((((((...(((....))).)))))))((....((((.((.....)).)))).---...))------)))).((((((((.........)))))))). ( -22.90, z-score = -1.20, R) >dp4.chr4_group3 259325 95 - 11692001 ------------GAAGGGGGCGUGGCGCAUGCAAAUGACACGCCUGAUGCUGCGUAAAUAAUUUUAGCGCU---CUCCG------CUGGCAUGAAAUGCAAAAUAAACAUUUUAUU ------------.....(((((((...(((....))).))))))).(((((((.((((....))))))((.---....)------).)))))((((((.........))))))... ( -23.80, z-score = 0.36, R) >droPer1.super_8 1299862 95 - 3966273 ------------GAAGGGGGCGUGGCGCAUGCAAAUGACACGCCUGAUGCUGCGUAAAUAAUUUUAGCGCU---CUCCG------CUGGCAUGAAAUGCAAAAUAAACAUUUUAUU ------------.....(((((((...(((....))).))))))).(((((((.((((....))))))((.---....)------).)))))((((((.........))))))... ( -23.80, z-score = 0.36, R) >droWil1.scaffold_180703 1069558 95 + 3946847 ------------UUGGGGGGCGUGGCACAUGCAAAUGACACGCCUGAUGCUGCGUAAAUAAUUUUAACGCUAGC--------CAACA-AAAUAAAAUGCAAAAUAAACAUUUUAUU ------------(((..(((((((...(((....))).)))))))...((((((((((....))).)))).)))--------...))-)(((((((((.........))))))))) ( -25.10, z-score = -2.09, R) >droVir3.scaffold_13246 1751799 100 - 2672878 -----------GUUUG-GGGUGUGGCGCAUGCAAAUGACACGCCCGAUGCUGCGUAAAUAAUUUUAGCGCUUGC--CAU--GCGCCACAAAUAAAAUGCAAAAUAAACAUUUUAUU -----------.....-...((((((((((((((.......((.....)).((((.((.....)).))))))).--)))--))))))))(((((((((.........))))))))) ( -31.10, z-score = -1.52, R) >droMoj3.scaffold_6500 4999864 107 + 32352404 ---------GUGGCAGGAGGCGUGGCGCAUGCAAAUGACACGCCUGAUGCUGCGUAAAUAAUUUUAGCGCUUGCCUCUGAUGCGCCAAAAAUAAAAUGCAAAAUAAACAUUUUAUU ---------(..(((..(((((((...(((....))).)))))))..)))..)........((((.((((...........)))).))))((((((((.........)))))))). ( -34.40, z-score = -2.06, R) >droGri2.scaffold_15252 16287582 113 + 17193109 UUAUAUUUUUGGUGCGUGGGCGUGGCGCAUGCAAAUGACACGCCCGAUGCUGCGUAAAUAAUUUUAGCGUUUGCGCCAU--GCGCCA-AAAUAAAAUGCAAAAUAAACAUUUUAUU ......((((((((((((((((((...(((....))).)))))))......((((((((.........)))))))).))--))))))-)))(((((((.........))))))).. ( -40.80, z-score = -2.76, R) >consensus ____________GCCGGGGGCGUGGCACAUGCAAAUGACACGCCUGAUGCUGCGUAAAUAAUUUUAGCGCU___CUCUC____UUCUGGCAUAAAAUGCAAAAUAAACAUUUUAUU .................(((((((...(((....))).)))))))......((((.((.....)).))))....................((((((((.........)))))))). (-21.72 = -21.08 + -0.64)
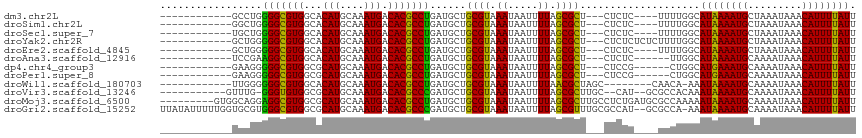
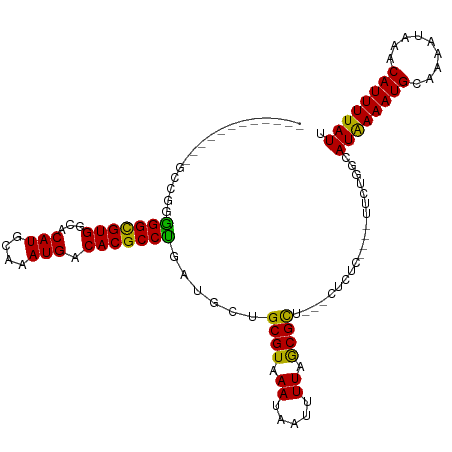

| Location | 17,086,798 – 17,086,895 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 82.90 |
| Shannon entropy | 0.34070 |
| G+C content | 0.41635 |
| Mean single sequence MFE | -26.91 |
| Consensus MFE | -17.99 |
| Energy contribution | -18.33 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.786540 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 17086798 97 - 23011544 AAUAAAAUGUUUAUUUAGCAUUUUAUGCCAAAA----GAGAG---AGCGCUAAAAUUAUUUACGCAGCAUCAGGCGUGUCAUUUGCAUGUGCCACGCCCCAGGC------------ .((((((((((.....))))))))))(((....----((...---.(((.((((....)))))))....)).((((((.(((......))).))))))...)))------------ ( -28.40, z-score = -2.70, R) >droSim1.chr2L 16770973 97 - 22036055 AAUAAAAUGUUUAUUUAGCAUUUUAUGCCAAAA----GAGAG---AGCGCUAAAAUUAUUUACGCAGCAUCAGGCGUGUCAUUUGCAUGUGCCACGCCCCAGCC------------ .((((((((((.....))))))))))((.....----((...---.(((.((((....)))))))....)).((((((.(((......))).))))))...)).------------ ( -25.00, z-score = -2.11, R) >droSec1.super_7 718537 97 - 3727775 AAUAAAAUGUUUAUUUAGCAUUUUAUGCCAAAA----GAGAG---AGCGCUAAAAUUAUUUACGCAGCAUCAGGCGUGUCAUUUGCAUGUGCCACGCCCCAGCA------------ .((((((((((.....))))))))))((.....----((...---.(((.((((....)))))))....)).((((((.(((......))).))))))...)).------------ ( -25.90, z-score = -2.13, R) >droYak2.chr2R 17126704 101 + 21139217 AAUAAAAUGUUUAUUUAGCAUUUUAUGCCAAAAGAGAGAGAG---AGCGCUAAAAUUAUUUACGCAGCAUCAGGCGUGUCAUUUGCAUGUGCCACGCCCCCAGC------------ .((((((((((.....))))))))))...........((...---.(((.((((....)))))))....)).((((((.(((......))).))))))......------------ ( -24.40, z-score = -1.71, R) >droEre2.scaffold_4845 15439093 97 + 22589142 AAUAAAAUGUUUAUUUAGCAUUUUAUGCCAAAA----GAGAG---AGCGCUAAAAUUAUUUACGCAGCAUCAGGCGUGUCAUUUGCAUGUGCCACGCCCCCAGC------------ .((((((((((.....)))))))))).......----((...---.(((.((((....)))))))....)).((((((.(((......))).))))))......------------ ( -24.40, z-score = -1.89, R) >droAna3.scaffold_12916 12418351 95 + 16180835 AAUAAAAUGUUUAUUUUGCAUUUUAUGCCAA------GAGAG---AGCGCUAAAAUUAUUUACGCAGCAUCAGGCGUGUCAUUUGCAUGUGCCACGCCUUCGGA------------ .(((((((((.......))))))))).((..------((...---.(((.((((....)))))))....))(((((((.(((......))).)))))))..)).------------ ( -26.50, z-score = -1.72, R) >dp4.chr4_group3 259325 95 + 11692001 AAUAAAAUGUUUAUUUUGCAUUUCAUGCCAG------CGGAG---AGCGCUAAAAUUAUUUACGCAGCAUCAGGCGUGUCAUUUGCAUGCGCCACGCCCCCUUC------------ ....((((((.......)))))).......(------((..(---(((((((((....)))).)).)).)).((((((((....).))))))).))).......------------ ( -19.80, z-score = 0.62, R) >droPer1.super_8 1299862 95 + 3966273 AAUAAAAUGUUUAUUUUGCAUUUCAUGCCAG------CGGAG---AGCGCUAAAAUUAUUUACGCAGCAUCAGGCGUGUCAUUUGCAUGCGCCACGCCCCCUUC------------ ....((((((.......)))))).......(------((..(---(((((((((....)))).)).)).)).((((((((....).))))))).))).......------------ ( -19.80, z-score = 0.62, R) >droWil1.scaffold_180703 1069558 95 - 3946847 AAUAAAAUGUUUAUUUUGCAUUUUAUUU-UGUUG--------GCUAGCGUUAAAAUUAUUUACGCAGCAUCAGGCGUGUCAUUUGCAUGUGCCACGCCCCCCAA------------ ((((((((((.......)))))))))).-((...--------(((.((((...........)))))))..))((((((.(((......))).))))))......------------ ( -27.20, z-score = -2.89, R) >droVir3.scaffold_13246 1751799 100 + 2672878 AAUAAAAUGUUUAUUUUGCAUUUUAUUUGUGGCGC--AUG--GCAAGCGCUAAAAUUAUUUACGCAGCAUCGGGCGUGUCAUUUGCAUGCGCCACACCC-CAAAC----------- ((((((((((.......))))))))))((((((((--(((--.((((((.((((....))))))).((.....)).......))))))))))))))...-.....----------- ( -34.10, z-score = -2.93, R) >droMoj3.scaffold_6500 4999864 107 - 32352404 AAUAAAAUGUUUAUUUUGCAUUUUAUUUUUGGCGCAUCAGAGGCAAGCGCUAAAAUUAUUUACGCAGCAUCAGGCGUGUCAUUUGCAUGCGCCACGCCUCCUGCCAC--------- ((((((((((.......))))))))))..(((((.....(((((..(((.((((....))))))).......((((((((....).)))))))..))))).))))).--------- ( -33.80, z-score = -2.72, R) >droGri2.scaffold_15252 16287582 113 - 17193109 AAUAAAAUGUUUAUUUUGCAUUUUAUUU-UGGCGC--AUGGCGCAAACGCUAAAAUUAUUUACGCAGCAUCGGGCGUGUCAUUUGCAUGCGCCACGCCCACGCACCAAAAAUAUAA ..((((((((.......))))))))(((-(((.((--.(((.((....((((((....)))).))......((((((((.....)))))).))..))))).)).))))))...... ( -33.60, z-score = -1.57, R) >consensus AAUAAAAUGUUUAUUUUGCAUUUUAUGCCAAAA____GAGAG___AGCGCUAAAAUUAUUUACGCAGCAUCAGGCGUGUCAUUUGCAUGUGCCACGCCCCCGGC____________ .(((((((((.......)))))))))....................(((.((((....))))))).......((((((.(((....)))...)))))).................. (-17.99 = -18.33 + 0.33)
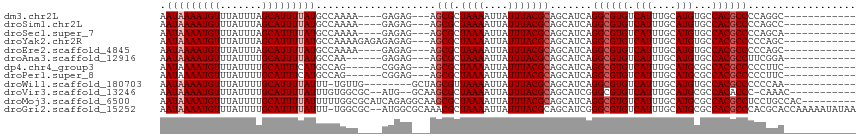
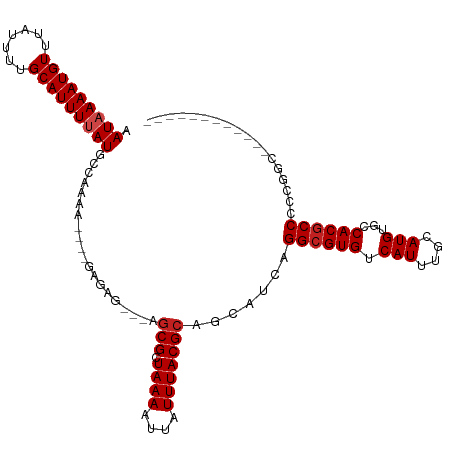
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:47:08 2011