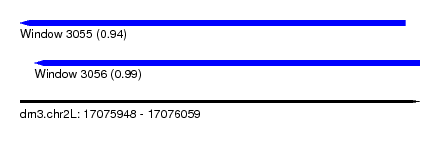
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 17,075,948 – 17,076,059 |
| Length | 111 |
| Max. P | 0.985009 |

| Location | 17,075,948 – 17,076,055 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 73.78 |
| Shannon entropy | 0.52718 |
| G+C content | 0.43095 |
| Mean single sequence MFE | -25.21 |
| Consensus MFE | -7.69 |
| Energy contribution | -7.80 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.82 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935130 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
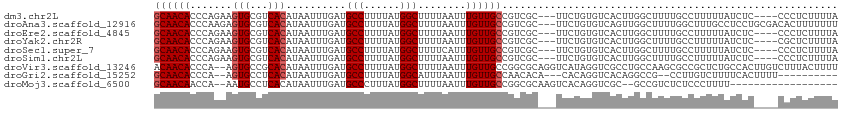
>dm3.chr2L 17075948 107 - 23011544 CACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGC---UUCUGUGUCACUUGGCUUUUGCCUUUUUAUCUC----CCCUCUUUUAUAUU .((.((((((((((.((.((((.((((.(((......)))..)))).)))))).)).)))---))))).)).....(((....)))..........----.............. ( -26.80, z-score = -4.68, R) >droAna3.scaffold_12916 12408543 111 + 16180835 CACCCAAGAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGC---UUCUGUGUCAGUUGGCUUUUGGCUUUGCCUCCUGCGACACUUUUUUUUAUU .....(((((.((((((.......)))))))))))..(((............)))(((((---......(((....)))....(((...)))....)))))............. ( -26.20, z-score = -1.52, R) >droEre2.scaffold_4845 15426540 107 + 22589142 CACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGC---UUCUGUGUCACUUGGCUUUUGCCUUUUUAUCUC----CCCUCUUUUAUAUU .((.((((((((((.((.((((.((((.(((......)))..)))).)))))).)).)))---))))).)).....(((....)))..........----.............. ( -26.80, z-score = -4.68, R) >droYak2.chr2R 17115251 107 + 21139217 CACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGC---UUCUGUGUCACUUGGCUUUUGCCUUUUUAUCUC----CGCUCUUUUAUAUU .((.((((((((((.((.((((.((((.(((......)))..)))).)))))).)).)))---))))).)).....(((....)))..........----.............. ( -26.80, z-score = -3.99, R) >droSim1.chr2L 16759940 107 - 22036055 CACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGC---UUCUGUGUCACUUGGCUUUUGCCUUUUUAUCUC----CCCUCUUUUAUAUG .((.((((((((((.((.((((.((((.(((......)))..)))).)))))).)).)))---))))).)).....(((....)))..........----.............. ( -26.80, z-score = -4.75, R) >droVir3.scaffold_13246 1741742 108 + 2672878 CACCCA--AGUGCCGCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGGCGCAGGUCAUAGGUCGCCUGCCAAGCGCCGCUCUGCCACUUGUCUUUACUUUU---- ....((--((((.((((.((((.((((.(((......)))..)))).)))))))((((((.(((...(((...))))))..))))))....).))))))...........---- ( -31.80, z-score = -1.43, R) >droGri2.scaffold_15252 16274327 93 - 17193109 CACCCA--AGUGCCUCACAUAAUUUGAUGCCUUUUAUGGCAUUUAAUUUGUUGCCAACACA---CACAGGUCACAGGCCGCCUUGUCUUUUCACUUUU---------------- ....((--((.((...(((.((((.((((((......)))))).)))))))..........---....(((.....))))))))).............---------------- ( -19.50, z-score = -1.40, R) >droMoj3.scaffold_6500 4987998 88 - 32352404 CAACCA--AAUGCCUCACAUAAUUUGAUGCCCUUUAUGGCUUUUAAUUUGUUGCCGGCGCAAGUCACAGGUCGC--GCCGUCUCUCCCUUUU---------------------- ((((((--(((..........)))))..(((......))).........)))).((((((............))--))))............---------------------- ( -17.00, z-score = -0.12, R) >consensus CACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGC___UUCUGUGUCACUUGGCUUUUGCCUUUUUAUCUC____CCCUCUUUUAUAUU .........(((((....((((.((((.(((......)))..)))).)))).((....))........)).)))..(((....)))............................ ( -7.69 = -7.80 + 0.11)

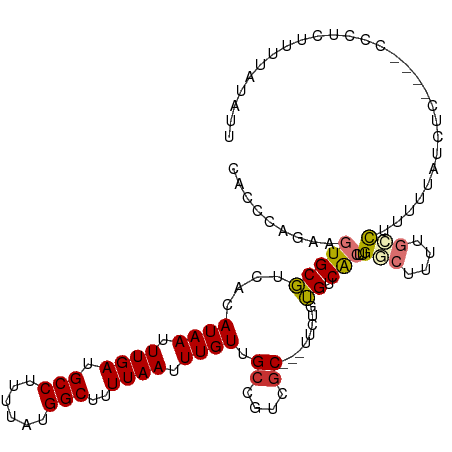
| Location | 17,075,952 – 17,076,059 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.18 |
| Shannon entropy | 0.47903 |
| G+C content | 0.43955 |
| Mean single sequence MFE | -26.66 |
| Consensus MFE | -10.80 |
| Energy contribution | -11.02 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.19 |
| SVM RNA-class probability | 0.985009 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 17075952 107 - 23011544 GCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGC---UUCUGUGUCACUUGGCUUUUGCCUUUUUAUCUC----CCCUCUUUUA .....((.((((((((((.((.((((.((((.(((......)))..)))).)))))).)).)))---))))).)).....(((....)))..........----.......... ( -26.80, z-score = -4.05, R) >droAna3.scaffold_12916 12408547 111 + 16180835 GCAACACCCAAGAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGC---UUCUGUGUCAGUUGGCUUUUGGCUUUGCCUCCUGCGACACUUUUUUU ((((((((.(((((.((((((.......)))))))))))..)).........)))))).(((((---......(((....)))....(((...)))....)))))......... ( -28.50, z-score = -1.58, R) >droEre2.scaffold_4845 15426544 107 + 22589142 GCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGC---UUCUGUGUCACUUGGCUUUUGCCUUUUUAUCUC----CCCUCUUUUA .....((.((((((((((.((.((((.((((.(((......)))..)))).)))))).)).)))---))))).)).....(((....)))..........----.......... ( -26.80, z-score = -4.05, R) >droYak2.chr2R 17115255 107 + 21139217 GCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGC---UUCUGUGUCACUUGGCUUUUGCCUUUUUAUCUC----CGCUCUUUUA ((...((.((((((((((.((.((((.((((.(((......)))..)))).)))))).)).)))---))))).)).....(((....)))..........----.))....... ( -28.80, z-score = -4.09, R) >droSec1.super_7 707751 107 - 3727775 GCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUCAUUUGUUGCCGUCGC---UUCUGUGUCACUUGGCUUUUGCCUUUUUAUCUC----CCCUCUUUUA .....((.((((((((((.((.((((..(((.(((......)))...))).)))))).)).)))---))))).)).....(((....)))..........----.......... ( -26.60, z-score = -3.95, R) >droSim1.chr2L 16759944 107 - 22036055 GCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGC---UUCUGUGUCACUUGGCUUUUGCCUUUUUAUCUC----CCCUCUUUUA .....((.((((((((((.((.((((.((((.(((......)))..)))).)))))).)).)))---))))).)).....(((....)))..........----.......... ( -26.80, z-score = -4.05, R) >droVir3.scaffold_13246 1741742 112 + 2672878 ACAACACCCA--AGUGCCGCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGGCGCAGGUCAUAGGUCGCCUGCCAAGCGCCGCUCUGCCACUUGUCUUUACUUUU ........((--((((.((((.((((.((((.(((......)))..)))).)))))))((((((.(((...(((...))))))..))))))....).))))))........... ( -31.80, z-score = -1.31, R) >droGri2.scaffold_15252 16274327 97 - 17193109 GCAACACCCA--AGUGCCUCACAUAAUUUGAUGCCUUUUAUGGCAUUUAAUUUGUUGCCAACACA---CACAGGUCACAGGCCG--CCUUGUCUUUUCACUUUU---------- ((((((....--.(((...)))..((((.((((((......)))))).))))))))))....(((---....(((.....))).--...)))............---------- ( -20.60, z-score = -1.41, R) >droMoj3.scaffold_6500 4987998 92 - 32352404 GCAACAACCA--AAUGCCUCACAUAAUUUGAUGCCCUUUAUGGCUUUUAAUUUGUUGCCGGCGCAAGUCACAGGUCGC--GCCGUCUCUCCCUUUU------------------ (((((((.((--(((..........)))))..(((......))).......)))))))((((((............))--))))............------------------ ( -23.20, z-score = -1.91, R) >consensus GCAACACCCAGAAGUGCGUCACAUAAUUUGAUGCCUUUUAUGGCUUUUAAUUUGUUGCCGUCGC___UUCUGUGUCACUUGGCUUUUGCCUUUUUAUCUC____CCCUCUUUUA ((((((.......(((...)))..........(((......)))........))))))........................................................ (-10.80 = -11.02 + 0.22)
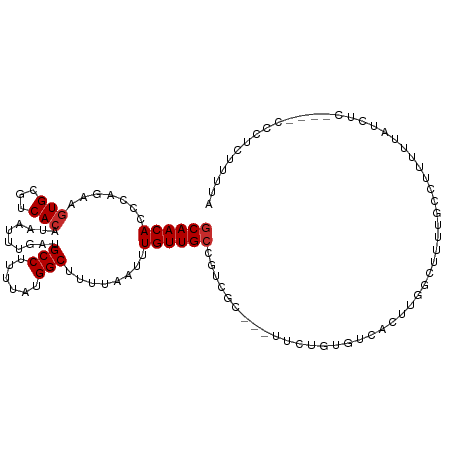
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:47:05 2011