| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,989,208 – 16,989,365 |
| Length | 157 |
| Max. P | 0.640114 |

| Location | 16,989,208 – 16,989,365 |
|---|---|
| Length | 157 |
| Sequences | 8 |
| Columns | 159 |
| Reading direction | forward |
| Mean pairwise identity | 70.67 |
| Shannon entropy | 0.56931 |
| G+C content | 0.40865 |
| Mean single sequence MFE | -41.11 |
| Consensus MFE | -10.12 |
| Energy contribution | -9.32 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.45 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640114 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
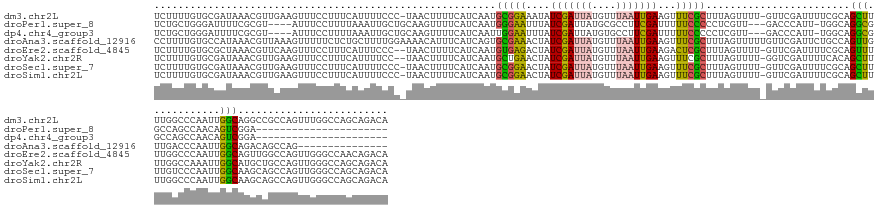
>dm3.chr2L 16989208 157 + 23011544 UCUUUUGUGCGAUAAACGUUGAAGUUUCCUUUCAUUUUCCC-UAACUUUUCAUCAAUGCGGAAAUAUCGAUUAUGUUUAAUUGAAGUUUCGCUUUAGUUUU-GUUCGAUUUUCGCAGCUUUUGGCCCAAUUGGCAGGCCGCCAGUUUGGCCAGCAGACA ...(((((((((((((.(((((((...........))))..-.))).))).)))..(((((((..(((((.((.....((((((((.....)))))))).)-).))))))))))))))...(((((.(((((((.....))))))).)))))))))).. ( -41.90, z-score = -2.08, R) >droPer1.super_8 1171557 129 - 3966273 UCUGCUGGGAUUUUCGCGU----AUUUCCUUUUAAAUUGCUGCAAGUUUUCAUCAAUGGGAAUUUAUCGAUUAUGCGCCUUCGAUUUUUCCCCCUCGUU---GACCCAUU-UGGCAGGCGGCCAGCCAACAGUCGGA---------------------- ((((((((((..(......----)..))).........(((((..(((....((((((((((...(((((..........)))))..)))))....)))---))......-.)))..))))).......))).))))---------------------- ( -30.30, z-score = 0.74, R) >dp4.chr4_group3 132638 129 - 11692001 UCUGCUGGGAUUUUCGCGU----AUUUCCUUUUAAAUUGCUGCAAGUUUUCAUCAAUUGGAAUUUAUCGAUUAUGUGCCUUCGAUUUUUCCCCCUCGUU---GACCCAUU-UGGCAGGCGGCCAGCCAACAGUCGGA---------------------- ((((((((((..(......----)..))).........(((((..(((....(((((.((((...(((((..........)))))..)))).....)))---))......-.)))..))))).......))).))))---------------------- ( -26.40, z-score = 1.44, R) >droAna3.scaffold_12916 12326253 144 - 16180835 CCUUUUGUGCCAUAAACGUUAAAGUUUUUCUCUGCUUUUGGAAAACAUUUCAUCAGUGCGAAACUAUCGAUUAUGUUUAAUUGAAGUUUCGCUUUAGUUUUUGUUCGAUUCUGCCAGUUGUUGACCCAAUUGGCAGACAGCCAG--------------- .....((.((((.((((......(((((((.........)))))))...........((((((((.(((((((....)))))))))))))))....)))).))......(((((((((((......)))))))))))..)))).--------------- ( -42.60, z-score = -4.48, R) >droEre2.scaffold_4845 15341928 156 - 22589142 UCUUUUGUGCGCUAAACGUUCAAGUUUCCUUUCAUUUCCC--UAACUUUUCAUCAAUGUGAGACUAUCGAUUAUGUUUAAUUGAAGACUCGCUUUAGUUUU-GUUCGAUUUUCGCAGUUUUUGGCCCAAUUGGCAGUUGGCCAGUUGGGCCAACAGACA .......((((..((.((..((((((..............--.)))...........(((((.((.(((((((....))))))))).))))).......))-)..)).))..))))((((((((((((((((((.....)))))))))))))).)))). ( -48.16, z-score = -4.42, R) >droYak2.chr2R 17021724 156 - 21139217 UCUUUUGUGCGAUAAACGUUGAAGUUUCCUUUCAUUUUCC--UAACUUUUCAUCAAUGCUGAACUAUCGAUUAUGUUUAAUUGAAGUUUCGCUUUAGUUUU-GGUCGAUUUUCACAGCUUUUGGCCAAAUUGGCAUGCUGCCAGUUGGGCCAGCAGACA .....(.(((((((((.(((((((...........)))).--.))).))).)))...((((....((((((((.....((((((((.....)))))))).)-))))))).....))))...(((((.(((((((.....))))))).)))))))).).. ( -42.20, z-score = -2.33, R) >droSec1.super_7 621321 157 + 3727775 UCUUUUGUGCGAUAAACGUUGAAGUUUCCUUUCAUUUUCCC-UAACUUUUCAUCAAUGCGGAACUAUCGAUUAUGUUUAAUUGAAGUUUCGCUUUAGUUUU-GUUCGAUUUUCGCAGCUUUUGUCCCAAUUGGCAAGCAGCCAGUUGGGCCAGCAGACA .......(((((.((.((.(((((.....))))).......-...............((((((((.(((((((....))))))))))))))).........-...)).)).)))))(((.(((.((((((((((.....)))))))))).))).)).). ( -45.10, z-score = -3.50, R) >droSim1.chr2L 16670083 157 + 22036055 UCUUUUGUGCGAUAAACGUUGAAGUUUCCUUUCAUUUUCCC-UAACUUUUCAUCAAUGCGGAACUAUCGAUUAUGUUUAAUUGAAGUUUCGCUUUAGUUUU-GUUCGAUUUUCGCAGCUUUUGGCCCAAUUGGCAAGCAGCCAGUUGGGCCAGCAGACA .......(((((.((.((.(((((.....))))).......-...............((((((((.(((((((....))))))))))))))).........-...)).)).)))))(((.((((((((((((((.....)))))))))))))).)).). ( -52.20, z-score = -4.95, R) >consensus UCUUUUGUGCGAUAAACGUUGAAGUUUCCUUUCAUUUUCCC_UAACUUUUCAUCAAUGCGAAACUAUCGAUUAUGUUUAAUUGAAGUUUCGCUUUAGUUUU_GUUCGAUUUUCGCAGCUUUUGGCCCAAUUGGCAGGCAGCCAGUUGGGCCAGCAGACA .........................................................(((((....(((((((....)))))))...)))))........................(((............)))......................... (-10.12 = -9.32 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:53 2011