| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,966,003 – 16,966,068 |
| Length | 65 |
| Max. P | 0.688032 |

| Location | 16,966,003 – 16,966,068 |
|---|---|
| Length | 65 |
| Sequences | 7 |
| Columns | 72 |
| Reading direction | forward |
| Mean pairwise identity | 65.07 |
| Shannon entropy | 0.63730 |
| G+C content | 0.42288 |
| Mean single sequence MFE | -12.21 |
| Consensus MFE | -5.68 |
| Energy contribution | -4.16 |
| Covariance contribution | -1.53 |
| Combinations/Pair | 2.00 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688032 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
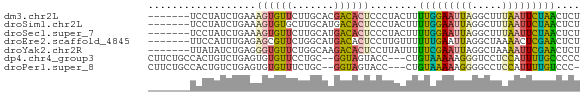
>dm3.chr2L 16966003 65 + 23011544 -------UCCUAUCUGAAAGUGUUCUUGCACGACACUCCCUACUUUUGGAAUUAGGCUUUAAUUCUAACUCU -------.((((((..((((((....((.....)).....))))))..))..))))................ ( -9.70, z-score = -1.01, R) >droSim1.chr2L 16647072 65 + 22036055 -------UCCUAUCUGAAAGUGUGCUUGCAUGACACUCCCUACUUUUGGAAUUAGGCUUUAAUUCUAACUCU -------.((((((..(((((((((......).))).....)))))..))..))))................ ( -11.40, z-score = -1.38, R) >droSec1.super_7 598109 65 + 3727775 -------UCCUAUCUGAAAGUGUUCUUGCAUGACACUCCCUACUUUUGGAAUUAGGCUUUAAUUCUAACUCU -------.((((((..((((((....((.....)).....))))))..))..))))................ ( -9.70, z-score = -0.89, R) >droEre2.scaffold_4845 15318899 65 - 22589142 -------UUCCAUUUGAGAGCGUUCUGGCAUGACACUCCUUGUUUUUUGAAUUAGGCUAAAACUCGAACUCU -------.....(((((((((.(.....((.((((.....))))...))....).)))....)))))).... ( -8.60, z-score = 0.86, R) >droYak2.chr2R 16997622 65 - 21139217 -------UUAUAUCUGAGGGUGUUCUGGCAAGACACUCCUUAUUUUUCGAAUUAGGCUAAAAUUCGAACUCU -------.......(((((((((.((....)))))).)))))...((((((((.......)))))))).... ( -16.20, z-score = -2.46, R) >dp4.chr4_group3 103327 67 - 11692001 CUUCUGCCACUGUCUGAGUGUGUUCCUGC--GGUAGUACC---CUGUAAAAAGGGUCCUCCAUUUUGCCCCC .....((((((.....)))).))......--((.((.(((---((......))))).))))........... ( -15.10, z-score = -1.34, R) >droPer1.super_8 1133017 66 - 3966273 CUUCUGCCACUGUCUGAGUGUGUUUCUGC--GGUAGUACC---CUGUAAAAAGGGGCCUCCAUUUUGUCCC- .....((((((.....)))).))......--((.((..((---((......))))..))))..........- ( -14.80, z-score = -0.44, R) >consensus _______UCCUAUCUGAGAGUGUUCUUGCAUGACACUCCCUACUUUUGGAAUUAGGCUUUAAUUCUAACUCU .............................................(((((((((.....))))))))).... ( -5.68 = -4.16 + -1.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:50 2011