
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,645,705 – 1,645,811 |
| Length | 106 |
| Max. P | 0.875544 |

| Location | 1,645,705 – 1,645,811 |
|---|---|
| Length | 106 |
| Sequences | 7 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.12 |
| Shannon entropy | 0.51929 |
| G+C content | 0.38560 |
| Mean single sequence MFE | -14.66 |
| Consensus MFE | -8.58 |
| Energy contribution | -10.26 |
| Covariance contribution | 1.68 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.875544 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1645705 106 - 23011544 UAACAACUU-AGCAGCCACAAUUGUCAAAGGUUUUCCACUUUGCUUUUCGGCUGCUUUCGUGUUUUC--GUUACCCCCUCCCC--CCUCUUUAUUUAUAUUUAUUUUAUGC .((((....-(((((((..((..(.(((((........))))))..)).)))))))....))))...--..............--.......................... ( -19.70, z-score = -3.61, R) >droSim1.chr2L 1679636 110 - 22036055 UAACAACUU-AGCAGCCACAAUUGUCAAAGGUUUUCCACUUUGCUUUUCGGCUGUUUUUGUUUUUCCCUUCCCUCCCCUCCCCGCUCCCUUUAUUUAUAUUUAUUUUAUGC .(((((...-(((((((..((..(.(((((........))))))..)).))))))).)))))................................................. ( -16.70, z-score = -3.01, R) >droSec1.super_14 1588398 99 - 2068291 UAAAAACUU-GGCAGCCACAAUUGUCAAAGGUUUUCCACUUUGCUUUUCGGCUGCUUUCGUUUUUUC--GCUUCCC---------CCCCUUUAUUUAUAUUUAUUUUAUGC .((((((..-(((((((..((..(.(((((........))))))..)).)))))))...))))))..--.......---------.......................... ( -19.90, z-score = -2.78, R) >droYak2.chr2L 1620851 106 - 22324452 UAACAACUU-GGCAGCCGCAAUUGUCAAAGGUUUUCCACUUUGCCUUUCGGCUGCUUUUGUUUUUCC--GC-UUCCUCCCCCCG-GCCCUUUAUUUAUAUUUAUUUUAUGC .(((((...-((((((((.((..(.(((((........))))))..)))))))))).)))))...((--(.-..........))-)......................... ( -22.50, z-score = -2.09, R) >droEre2.scaffold_4929 1695059 107 - 26641161 UAACAACUU-GGCAGCCGCAAUUGUCAAAGGUUUUCCACUUUGCUUUUCGGCUGCUUGUGUUUUUCC--GCUUUUCCCUCCCCG-GCCCUUUAUUUAUAUUUAUUUUAUGC .((((....-((((((((.((..(.(((((........))))))..))))))))))..))))...((--(............))-)......................... ( -23.20, z-score = -2.08, R) >dp4.chr4_group2 548938 83 - 1235136 --ACAAUUUCACCAGCCACAAUUGUCCAUGAUGUUUUCCCCAGUUUUCU--UUGCUCUUUUUCUG--------------------CCUUUUUAUUUAU----UUUUUAUGC --((((((...........))))))........................--..............--------------------.............----......... ( -0.30, z-score = 2.19, R) >droPer1.super_8 624212 83 + 3966273 --ACAAUUUCACCAGCCACAAUUGUCCAUGAUGUUUUCCCCAGUUUUCU--UUGCUCUUUUUCUG--------------------CCUUUUUAUUUAU----UUUUUAUGC --((((((...........))))))........................--..............--------------------.............----......... ( -0.30, z-score = 2.19, R) >consensus UAACAACUU_AGCAGCCACAAUUGUCAAAGGUUUUCCACUUUGCUUUUCGGCUGCUUUUGUUUUUCC__GC_UUCC_C_CCCC__CCCCUUUAUUUAUAUUUAUUUUAUGC ....(((...((((((((.((..(.(((((........)))))).)).))))))))...)))................................................. ( -8.58 = -10.26 + 1.68)
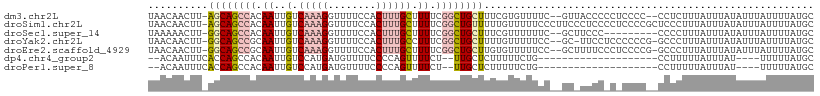

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:51 2011