| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,964,992 – 16,965,073 |
| Length | 81 |
| Max. P | 0.742123 |

| Location | 16,964,992 – 16,965,073 |
|---|---|
| Length | 81 |
| Sequences | 9 |
| Columns | 82 |
| Reading direction | reverse |
| Mean pairwise identity | 76.99 |
| Shannon entropy | 0.49583 |
| G+C content | 0.41743 |
| Mean single sequence MFE | -18.57 |
| Consensus MFE | -10.69 |
| Energy contribution | -11.23 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.742123 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16964992 81 - 23011544 AGGGUAUCAAGAUGAA-GAAAUGUAAGACUAACAUCUCCAGCAUCCAUUUGCUGGAAUGAAAAAUGGCUCUAGCAAACAACU (((((.....((((..-...............))))(((((((......)))))))..........)))))........... ( -16.93, z-score = -1.09, R) >droSim1.chr2L 16645906 81 - 22036055 AGGGUAUCAAGAUGGA-GAAAUGUAAGACUAACAUCUCCAGCAGCCAUUUGCUGGAAUGAAAAAUGGCUCUGGCAAACAACU ..(((.......((((-((..(((.......)))))))))((((((((((...........))))))))...)).....))) ( -18.30, z-score = -0.69, R) >droSec1.super_7 597135 75 - 3727775 AGGGUAUCAAGAUGGA-GAAAUG------UAACAUCUCCAGCAGACAUUUGCUGGAAUGAAAAAUGGCUCUCGCAAACAACU .(((..(((.((((..-......------...))))((((((((....))))))))........)))..))).......... ( -18.10, z-score = -1.61, R) >droYak2.chr2R 16996556 81 + 21139217 AGGGUAUCAAGAUGGA-GAAAUGUAAGACUAACAUCUCCAGCAGCCAUUUGCUGGAAUGAAAAAUGGCUCUAGCAAACAACU ..(((.......((((-((..(((.......)))))))))...))).(((((((((............)))))))))..... ( -19.10, z-score = -1.28, R) >droEre2.scaffold_4845 15317912 81 + 22589142 AGGGUAUCAAGAUGAA-GAAAUAUAAGACUAACAUCUACAGCAGCCAUUUGCUGGAAUGAAAAAUGGCUCCGGCAAACAACU ..(((....(((((..-...............)))))......))).(((((((((............)))))))))..... ( -14.93, z-score = -0.70, R) >droAna3.scaffold_12916 12304099 82 + 16180835 AGGGUAUCCCAAAAACCAAGACCUAUGUCUUAAAUCUCCAGCUGCCACUUGCUGGGAUGAAAAAUUGCUCUGGCAAACAACU .(((...))).......(((((....)))))..(((.(((((........))))))))......((((....))))...... ( -20.10, z-score = -1.56, R) >dp4.chr4_group3 101891 82 + 11692001 AGGGCAUCGAGAUGGGUGAAAAGUAAGACUAACAUCUCCAGCAACCAUUUGCUGGGAUGAAAAAUUGCUUUGGCAAACAAGU ............((..((.(((((((......((((.(((((((....))))))))))).....)))))))..))..))... ( -23.90, z-score = -2.36, R) >droPer1.super_8 1131574 82 + 3966273 AGGGCAUCGAGAUGGGUGAAAAGUAAGACUAACAUCUCCAGCAACCAUUUGCUGGGAUGAAAAAUUGCUUUGGCAAACAAGU ............((..((.(((((((......((((.(((((((....))))))))))).....)))))))..))..))... ( -23.90, z-score = -2.36, R) >droWil1.scaffold_180703 2726964 74 + 3946847 ----UGCCUAGAUAAA-GCGAAAUAA---CCGUGCCCCAGCAAACCAUUUGGUCAAAUGAAAAAUUGCAUCUGCCAGGUGCU ----(((.........-(((......---.)))......)))...(((((((.((.(((........))).))))))))).. ( -11.86, z-score = 0.60, R) >consensus AGGGUAUCAAGAUGGA_GAAAUGUAAGACUAACAUCUCCAGCAGCCAUUUGCUGGAAUGAAAAAUGGCUCUGGCAAACAACU (((((.....((((..................))))((((((((....))))))))..........)))))........... (-10.69 = -11.23 + 0.54)


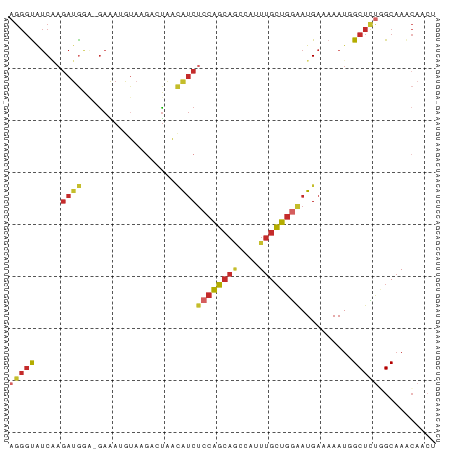
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:50 2011