| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,941,188 – 16,941,281 |
| Length | 93 |
| Max. P | 0.727967 |

| Location | 16,941,188 – 16,941,281 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 68.13 |
| Shannon entropy | 0.60289 |
| G+C content | 0.45197 |
| Mean single sequence MFE | -25.28 |
| Consensus MFE | -11.69 |
| Energy contribution | -11.72 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727967 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
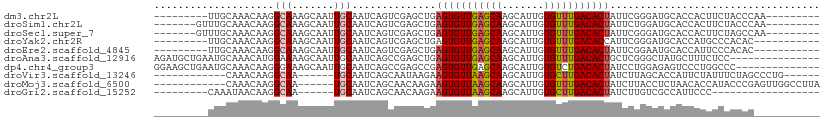
>dm3.chr2L 16941188 93 - 23011544 ---------UUGCAAACAAGGCAAAGCAAUUGCAAUCAGUCGAGCUGAGUGUUGAGCAAGCAUUGUGUUUGACACUAUUCGGGAUGCACCACUUCUACCCAA--------- ---------.((((..(...((((.....)))).......((((...(((((..((((.......))))..))))).)))))..))))..............--------- ( -22.50, z-score = 0.22, R) >droSim1.chr2L 16630174 95 - 22036055 -------GUUUGCAAACAAGGCAAAGCAAUUGCAAUCAGUCGAGCUGAGUGUUGAGCAAGCAUUGUGUUUGACACUAUUCUGGAUGCACCACUUCUACCCAA--------- -------...(((.......((((.....)))).....(((.((...(((((..((((.......))))..)))))...)).))))))..............--------- ( -22.50, z-score = 0.48, R) >droSec1.super_7 578278 95 - 3727775 -------GUUUGCAAACAAGGCAAAGCAAUUGCAAUCAGUCGAGCUGAGUGUUGAGCAAGCAUUGUGUUUGACACUAUUCGGGAUGCACCACUUCUAGCCAA--------- -------............(((.(((....((((.((...((((...(((((..((((.......))))..))))).)))).))))))...)))...)))..--------- ( -24.00, z-score = 0.40, R) >droYak2.chr2R 16976158 91 + 21139217 ---------UUGCAAACAAGGCAAAGCAAUUGCAAUCAGUCGAGCUGAGUGUUGAGCAAGCAUUGUGUUUGACACCAUUCGGGAUGCACCAUGCCCACAC----------- ---------((((.......)))).(((..((((.((...((((.((.((((..((((.......))))..)))))))))).))))))...)))......----------- ( -24.30, z-score = -0.07, R) >droEre2.scaffold_4845 15298965 91 + 22589142 ---------UUGCAAACAAGGCAAAGCAAUUGCAAUCAGUCGAGCUGAGUGUUGAGCAAGCAUUGUGUUUGACACUAUUCGGAAUGCACCAUUCCCACAC----------- ---------((((((.....((...))..))))))((((.....))))((((..((((.......))))..)))).....((((((...)))))).....----------- ( -23.30, z-score = -0.54, R) >droAna3.scaffold_12916 12288111 96 + 16180835 AGAUGCUGAAUGCAAACAUGGAAAAGCAAUUGCAAUCAGCCGAGCUGAGUGUUGAGCAAGCAUUGUGUUUGACACUGCUCGGGCUAUGCUUUCUCC--------------- ...(((.....))).....(((((((((...((......((((((..(((((..((((.......))))..)))))))))))))..)))))).)))--------------- ( -32.80, z-score = -1.80, R) >dp4.chr4_group3 80022 97 + 11692001 GGAAGCUGAAUGCAAACAAGGGAAAGCAAUUGCAAUCAGCCGAGCCGAGUGUUGAGCAAGCAUUGUGUCUGACACUAUCCUGGAGAGUCCCUGGCCC-------------- ((..(((((.(((((..............))))).)))))...((((((((((..(((.......)))..)))))).....((.....)).))))))-------------- ( -25.54, z-score = 0.35, R) >droVir3.scaffold_13246 1580161 87 + 2672878 ------------CAAACAAGGCAA------UGCAAUCAGCAAUAAGAAGUGUUAAGCAAGCAUUGUGCUUGACACUAUCUUAGCACCAUUCUAUUUCUAGCCCUG------ ------------....((.(((..------(((.....))).((((((((((((((((.......))))))))))).))))).................))).))------ ( -25.70, z-score = -3.35, R) >droMoj3.scaffold_6500 4814887 93 - 32352404 ------------CAAACAAGGCAA------UGCAAUCAGCAACAAGAAGUGUUAAGCAAGCAUUGUGUUUGACACUAUCUUACCUCUAACACCAUACCCGAGUUGGCCUUA ------------.....(((((..------(((.....)))..(((((((((((((((.......))))))))))).))))........................))))). ( -23.70, z-score = -2.29, R) >droGri2.scaffold_15252 16126157 78 - 17193109 ---------CAAAUAACAAGGCAA------UGCAAUCAGCAACAAGAAGUGUUAAGCAAGCAUUGUGCUUGACACUAUCUUGUCGCCAUUCCC------------------ ---------..........(((..------(((.....)))(((((((((((((((((.......))))))))))).)))))).)))......------------------ ( -28.50, z-score = -5.42, R) >consensus _________UUGCAAACAAGGCAAAGCAAUUGCAAUCAGCCGAGCUGAGUGUUGAGCAAGCAUUGUGUUUGACACUAUUCGGGAUGCACCACUUCCACCC___________ ....................(((.......)))..............(((((((((((.......)))))))))))................................... (-11.69 = -11.72 + 0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:47 2011