
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,920,904 – 16,920,976 |
| Length | 72 |
| Max. P | 0.567142 |

| Location | 16,920,904 – 16,920,976 |
|---|---|
| Length | 72 |
| Sequences | 12 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 77.83 |
| Shannon entropy | 0.45076 |
| G+C content | 0.45535 |
| Mean single sequence MFE | -18.64 |
| Consensus MFE | -11.37 |
| Energy contribution | -11.07 |
| Covariance contribution | -0.29 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567142 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 16920904 72 + 23011544 GCUGAGGAAGUGCACACACAAG------AAUUUCGCCACUCGACUGUGGCAGAGUAAUUUCAUAAUUAUGCAAGCACA .........((((.........------......(((((......))))).(.((((((....)))))).)..)))). ( -18.10, z-score = -1.35, R) >droSim1.chr2L 16609682 72 + 22036055 GCUGAGGAAGUGCACACACAAG------AAUUUCGCCACUCGACUGUGGCAGAGUAAUUUCAUAAUUAUGCAAGCACA .........((((.........------......(((((......))))).(.((((((....)))))).)..)))). ( -18.10, z-score = -1.35, R) >droSec1.super_7 558216 72 + 3727775 GCUGAGGAAGUGCACACACAAG------AAUUUCGCCACUCGACUGCGGCAGAGUAAUUUCAUAAUUAUGCAAGCACA .........((((.........------......(((.(......).))).(.((((((....)))))).)..)))). ( -14.20, z-score = 0.03, R) >droYak2.chr2R 16955570 72 - 21139217 GCUGAGGAAGUGCACAUACAAG------UAUUUCGCCACUCGACUGUGGCAGAGUAAUUUCAUAAUUAUGCAAGCACA (((...(((((((........)------))))))(((((......))))).(.((((((....)))))).).)))... ( -20.30, z-score = -2.06, R) >droEre2.scaffold_4845 15278691 72 - 22589142 GCUGAGGAAGUGCACACACAAG------AAUUUCGCCACUCGACUGUGGCAGAGUAAUUUCAUAAUUAUGCAAGCACA .........((((.........------......(((((......))))).(.((((((....)))))).)..)))). ( -18.10, z-score = -1.35, R) >droAna3.scaffold_12916 12268521 72 - 16180835 ACUGUGGAAGUGCGCACACAAG------AAUUUCGCCACUCGACUGUGGCAGAGUAAUUUCAUAAUUAUGCAAGCACA .........((((((.......------......(((((......)))))...((((((....))))))))..)))). ( -18.20, z-score = -0.85, R) >dp4.chr4_group3 56249 78 - 11692001 GCAGAGGAAGUGCACACAGAGAGCGAACAGUUUCGCCACUCGGCUGUGGCAGAGUAAUUUCAUAAUUAUGCAGGCACA .........((((.(((((.(((((((....)))))...))..)))))(((...(((((....))))))))..)))). ( -21.40, z-score = -1.21, R) >droPer1.super_8 1085452 78 - 3966273 GCCGAGGAAGUGCACACAGAGAGCGAACAGUUUCGCCACUCGGCUGUGGCAGAGUAAUUUCAUAAUUAUGCAGGCACA ((((((...((....))...(.(((((....)))))).)))))).(((.(...((((((....))))))...).))). ( -24.10, z-score = -1.86, R) >droWil1.scaffold_180703 2640493 58 - 3946847 -CUAAGGAAGUGC------------------UUUGGCGCAGUUCU-UGCCAAAGUAAUUCCAUAAUUAUGCAAACGCA -....((((.(((------------------(((((((.......-)))))))))).)))).......(((....))) ( -19.90, z-score = -3.57, R) >droMoj3.scaffold_6500 27567932 62 - 32352404 GCUAAGGAAGUGCGCGUUGU------------UUGCCACUUGGC---GGCA-AGUAAUUUUAUAAUUAUGCAAGCGCA ..........(((((.((((------------(((((.......---))))-)((((((....))))))))))))))) ( -17.10, z-score = -0.08, R) >droVir3.scaffold_13246 1127223 62 + 2672878 GCUAAGGAAGUGCGCGUUGU------------UUGCCACUUGGC---GGCA-AGUAAUUUUAUAAUUAUGCAAGCGCA ..........(((((.((((------------(((((.......---))))-)((((((....))))))))))))))) ( -17.10, z-score = -0.08, R) >droGri2.scaffold_15252 1093891 62 - 17193109 GCCAAGGAAGUGCGCGUUGU------------UUGCCACUUGGC---GGCA-AGUAAUUUUAUAAUUAUGCAAGCGCA ..........(((((.((((------------(((((.......---))))-)((((((....))))))))))))))) ( -17.10, z-score = 0.00, R) >consensus GCUGAGGAAGUGCACACACAAG______AAUUUCGCCACUCGACUGUGGCAGAGUAAUUUCAUAAUUAUGCAAGCACA .........((((.....................(((..........)))...((((((....))))))....)))). (-11.37 = -11.07 + -0.29)
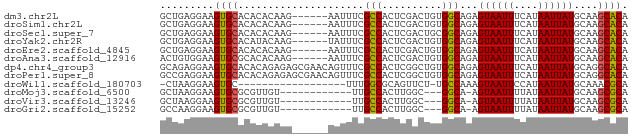

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:45 2011