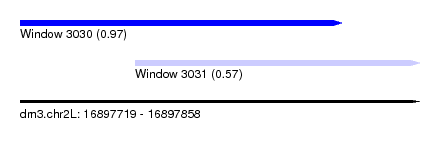
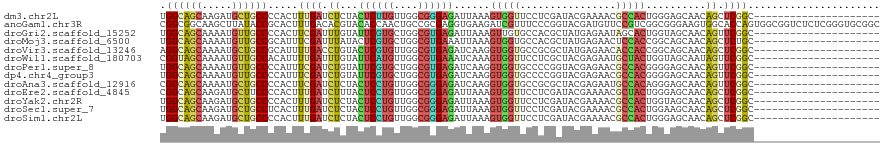
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,897,719 – 16,897,858 |
| Length | 139 |
| Max. P | 0.970995 |

| Location | 16,897,719 – 16,897,831 |
|---|---|
| Length | 112 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.94 |
| Shannon entropy | 0.42197 |
| G+C content | 0.51342 |
| Mean single sequence MFE | -39.59 |
| Consensus MFE | -22.87 |
| Energy contribution | -21.32 |
| Covariance contribution | -1.54 |
| Combinations/Pair | 1.66 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970995 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
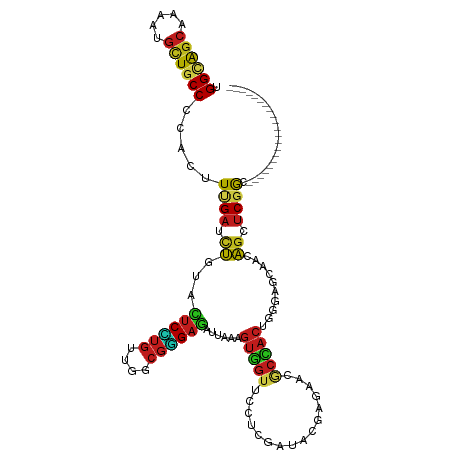
>dm3.chr2L 16897719 112 + 23011544 CACUCUUCUCGGAGAACGAAGACGGUGGAUUGACCUAUAUUGGCAGCAAGAUGCUGCCCCACUUUGAUCUCUACUCUUGUUGGCGGGAGAUUAAAGUGGUUCCUCGAUACGA-------- ((((.(..(((.....)))..).)))).(((((........((((((.....))))))((((((((((((((..((.....))..))))))))))))))....)))))....-------- ( -41.70, z-score = -3.56, R) >anoGam1.chr3R 49933760 120 - 53272125 CACUGUUUCAGCAAAACGAGGACGGCGGGCUGGUGUACAUCGGCGGCAAGCUUAUACCGCACUUUGACACGUACACCAACUGCCGCGAGGUGAAGAUCGUUUCCCGGUACGAUGUUCCGU ..((((((.....)))).))(.((((((..((((((((.(((((((..........))))....)))...)))))))).)))))))...(.(((.(((((........))))).)))).. ( -36.90, z-score = 0.37, R) >droGri2.scaffold_15252 16078222 112 + 17193109 CGCUCUUCCACGAGAACGAGGACGGUGGCCUAACCUAUAUUGGCAGCAAAAUGUUGCCCCACUUCGAUUUGUAUUCGUGCUGGCGUGAGAUUAAAGUUGUGCCACGCUAUGA-------- .((.....((((((.(((((...(((((.............((((((.....)))))))))))....))))).)))))).(((((..(........)..))))).)).....-------- ( -33.01, z-score = -1.00, R) >droMoj3.scaffold_6500 4763637 112 + 32352404 CGCUCUUCCAUGAGAACGAAGACGGUGGCCUCACCUACAUUGGCAGCAAAAUGUUGCCGCAUUUCGAUUUAUACUCGUGCUGGCGUGAAAUUAAAGUGGUGCCACGCUAUGA-------- ((.(((((.........)))))))(((((.((((.......((((((.....))))))((....(((.......))).....))...........)))).))))).......-------- ( -31.50, z-score = -0.43, R) >droVir3.scaffold_13246 1525056 112 - 2672878 CGCUCUUCCACGAGAACGAGGAUGGCGGCCUGACCUAUAUAGGCAGCAAAAUGCUGCCGCAUUUUGACCUGUACUCGUGUUGGCGUGAGAUCAAGGUGGUGCCGCGCUAUGA-------- .((...(((.((....)).)))..(((((.(.((((.((((((...((((((((....)))))))).))))))((((((....))))))....)))).).))))))).....-------- ( -41.50, z-score = -1.62, R) >droWil1.scaffold_180703 2619421 112 - 3946847 CUCUUUUUCACGAGAACGAAGAUGGUGGCCUGACUUAUAUCGGUAGCAAAAUGUUGCCACAUUUUGAUUUGUAUUCAUGUUGGCGUGAAAUCAAAGUGGUUCCUCGCUACGA-------- .((((((((....))).)))))..(((((............((((((.....)))))).((((((((((.....(((((....))))))))))))))).......)))))..-------- ( -31.00, z-score = -1.61, R) >droPer1.super_8 1065911 112 - 3966273 CGCUCUUCAGCGAGAACGAAGAUGGAGGUCUAACCUACAUUGGCAGCAAAAUGUUGCCCCAUUUCGAUCUGUAUUCGUGCUGGCGUGAGAUCAAGGUGGUGCCCCGGUACGA-------- ((((....))))(((.(((.((((((((.....))).....((((((.....)))))))))))))).)))....(((((((((.(((..((....))..))).)))))))))-------- ( -40.20, z-score = -2.04, R) >dp4.chr4_group3 36741 112 - 11692001 CGCUCUUCAGCGAGAACGAAGAUGGAGGUCUAACCUACAUUGGCAGCAAAAUGUUGCCCCAUUUCGAUCUGUAUUCGUGCUGGCGUGAGAUCAAGGUGGUGCCCCGGUACGA-------- ((((....))))(((.(((.((((((((.....))).....((((((.....)))))))))))))).)))....(((((((((.(((..((....))..))).)))))))))-------- ( -40.20, z-score = -2.04, R) >droAna3.scaffold_12916 12247443 112 - 16180835 CGCUCUUCCACGAGAACGAGGACGGCGGCCUCACCUACAUCGGCAGCAAAAUGCUGCCCCACUUCGAUCUCUACUCCUGUUGGCGGGAGAUCAAGGUGGUGCCGCGCUACGA-------- .((.(.(((.((....)).))).)(((((.((((((.....((((((.....)))))).......(((((((...((....))..))))))).)))))).))))))).....-------- ( -49.40, z-score = -3.62, R) >droEre2.scaffold_4845 15254762 112 - 22589142 CCCUCUUCUCGGAGAACGAAGACGGCGGAUUAACCUAUAUCGGCAGCAAGAUGCUUCCCCACUUUGAUCUUUACUCCUGUUGGCGGGAGAUUAAAGUGGUUCCUCGAUACGA-------- ((((.(..(((.....)))..).)).)).........((((((.(((.....))).))((((((((((((((...((....))..))))))))))))))......))))...-------- ( -33.50, z-score = -0.93, R) >droYak2.chr2R 16932660 112 - 21139217 CACUCUUCUCGGAGAACGAAGACGGUGGAUUGACCUACAUUGGCAGCAAGAUGCUGCCCCACUUUGAUCUCUACUCCUGUUGGCGGGAGAUUAAAGUGGUUCCUCGAUACGA-------- ((((.(..(((.....)))..).)))).(((((........((((((.....))))))((((((((((((((...((....))..))))))))))))))....)))))....-------- ( -44.00, z-score = -3.96, R) >droSec1.super_7 534865 112 + 3727775 CACUCUUCUCGGAGAACGAAGACGGUGGAUUGACCUAUAUUGGCAGCAAGAUGCUGCCUCACUUUGAUCUCUACUCCUGUUGGCGGGAGAUUAAAGUGGUUCCUCGAUACGA-------- ........(((..((((......(((......)))......((((((.....)))))).(((((((((((((...((....))..)))))))))))))))))..))).....-------- ( -42.50, z-score = -3.72, R) >droSim1.chr2L 16583922 112 + 22036055 CACUGUUCUCGGAGAACGAAGACGGUGGAUUGACCUAUAUUGGCAGCAAGAUGCUGCCCCACUUUGAUCUCUACUCCUGUUGGCGGGAGAUUAAAGUGGUUCCUCGAUACGA-------- ((((((..(((.....)))..)))))).(((((........((((((.....))))))((((((((((((((...((....))..))))))))))))))....)))))....-------- ( -49.30, z-score = -5.62, R) >consensus CGCUCUUCCACGAGAACGAAGACGGUGGACUGACCUAUAUUGGCAGCAAAAUGCUGCCCCACUUUGAUCUGUACUCCUGUUGGCGGGAGAUUAAAGUGGUUCCUCGAUACGA________ ...(((((.........)))))..(.(((............((((((.....)))))).(((((.((((.....(((((....))))))))).)))))..))).)............... (-22.87 = -21.32 + -1.54)
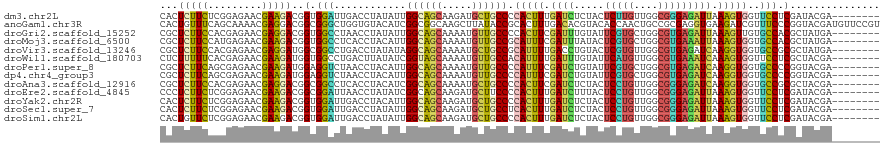
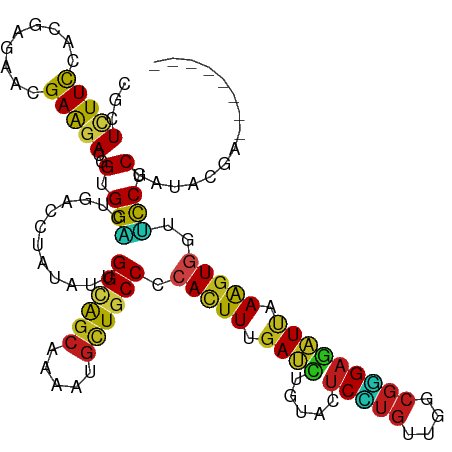
| Location | 16,897,759 – 16,897,858 |
|---|---|
| Length | 99 |
| Sequences | 13 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.70 |
| Shannon entropy | 0.43753 |
| G+C content | 0.54147 |
| Mean single sequence MFE | -35.85 |
| Consensus MFE | -18.51 |
| Energy contribution | -18.01 |
| Covariance contribution | -0.49 |
| Combinations/Pair | 1.65 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.570809 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16897759 99 + 23011544 UGGCAGCAAGAUGCUGCCCCACUUUGAUCUCUACUCUUGUUGGCGGGAGAUUAAAGUGGUUCCUCGAUACGAAAACGCCACUGGGAGCAACAGCUCGGC--------------------- .((((((.....))))))((((((((((((((..((.....))..)))))))))))))).................(((.....((((....)))))))--------------------- ( -38.40, z-score = -2.92, R) >anoGam1.chr3R 49933800 120 - 53272125 CGGCGGCAAGCUUAUACCGCACUUUGACACGUACACCAACUGCCGCGAGGUGAAGAUCGUUUCCCGGUACGAUGUUCCGUCGGCGGGAAGUGGCACCAGUGGCGGUCUCUCGGGUGCGGC ..((((..........)))).........(((((.((.((((((((..((((....((((((((((...(((((...))))).)))))))))))))).)))))))).....))))))).. ( -47.00, z-score = -0.33, R) >droGri2.scaffold_15252 16078262 99 + 17193109 UGGCAGCAAAAUGUUGCCCCACUUCGAUUUGUAUUCGUGCUGGCGUGAGAUUAAAGUUGUGCCACGCUAUGAGAAUAGCACUGGUAGCAACAGUUCGGC--------------------- .((((((.....)))))).....((((.((((........(((((..(........)..))))).(((((.((.......)).))))).)))).)))).--------------------- ( -29.80, z-score = -0.70, R) >droMoj3.scaffold_6500 4763677 99 + 32352404 UGGCAGCAAAAUGUUGCCGCAUUUCGAUUUAUACUCGUGCUGGCGUGAAAUUAAAGUGGUGCCACGCUAUGAGAACUCCACCGGCAGCAACAGCUCUGC--------------------- .((((((.....))))))((....(((.......)))((((((.(.((..(((.((((......)))).)))....))).))))))))...........--------------------- ( -27.80, z-score = 0.11, R) >droVir3.scaffold_13246 1525096 99 - 2672878 AGGCAGCAAAAUGCUGCCGCAUUUUGACCUGUACUCGUGUUGGCGUGAGAUCAAGGUGGUGCCGCGCUAUGAGAACACCACCGGCAGCAACAGCUCGGC--------------------- .((((((.....)))))).....((((.((((.....(((((.(.((....)).(((((((...(.......)..)))))))).))))))))).)))).--------------------- ( -35.30, z-score = -0.34, R) >droWil1.scaffold_180703 2619461 99 - 3946847 CGGUAGCAAAAUGUUGCCACAUUUUGAUUUGUAUUCAUGUUGGCGUGAAAUCAAAGUGGUUCCUCGCUACGAGAAUGCUACUGGUAGCAAUAGUUCGGC--------------------- .((((((.....)))))).((((((((((.....(((((....))))))))))))))).......(((((.((.......)).)))))...........--------------------- ( -30.50, z-score = -2.40, R) >droPer1.super_8 1065951 99 - 3966273 UGGCAGCAAAAUGUUGCCCCAUUUCGAUCUGUAUUCGUGCUGGCGUGAGAUCAAGGUGGUGCCCCGGUACGAGAACGCCACGGGGAGCAACAGUUCGGC--------------------- .((((((.....)))))).....((((.((((.((((((((((.(((..((....))..))).))))))))))...((........)).)))).)))).--------------------- ( -34.30, z-score = -0.44, R) >dp4.chr4_group3 36781 99 - 11692001 UGGCAGCAAAAUGUUGCCCCAUUUCGAUCUGUAUUCGUGCUGGCGUGAGAUCAAGGUGGUGCCCCGGUACGAGAACGCCACGGGGAGCAACAGUUCGGC--------------------- .((((((.....)))))).....((((.((((.((((((((((.(((..((....))..))).))))))))))...((........)).)))).)))).--------------------- ( -34.30, z-score = -0.44, R) >droAna3.scaffold_12916 12247483 99 - 16180835 CGGCAGCAAAAUGCUGCCCCACUUCGAUCUCUACUCCUGUUGGCGGGAGAUCAAGGUGGUGCCGCGCUACGAGAAUGCCACAGGGAGCAACAGUUCGGC--------------------- .((((((.....))))))((((((.(((((((...((....))..))))))).)))))).((((.((.........))).....((((....)))))))--------------------- ( -39.70, z-score = -1.84, R) >droEre2.scaffold_4845 15254802 99 - 22589142 CGGCAGCAAGAUGCUUCCCCACUUUGAUCUUUACUCCUGUUGGCGGGAGAUUAAAGUGGUUCCUCGAUACGAAAACGCUACUGGGAGCAACAGCUCGGC--------------------- .(.((((....(((((((((((((((((((((...((....))..))))))))))))))....(((...)))..........)))))))...))).).)--------------------- ( -32.10, z-score = -1.43, R) >droYak2.chr2R 16932700 99 - 21139217 UGGCAGCAAGAUGCUGCCCCACUUUGAUCUCUACUCCUGUUGGCGGGAGAUUAAAGUGGUUCCUCGAUACGAAAACGCCACUGGUAGCAACAGCUCGGC--------------------- .((((((.....))))))((((((((((((((...((....))..)))))))))))))).................(((.(((.......)))...)))--------------------- ( -38.90, z-score = -3.24, R) >droSec1.super_7 534905 99 + 3727775 UGGCAGCAAGAUGCUGCCUCACUUUGAUCUCUACUCCUGUUGGCGGGAGAUUAAAGUGGUUCCUCGAUACGAAAACGCCACUGGAAGCAACAGCUCGGC--------------------- .((((((.....)))))).(((((((((((((...((....))..)))))))))))))(((..(((...))).)))(((.(((.......)))...)))--------------------- ( -37.20, z-score = -2.97, R) >droSim1.chr2L 16583962 99 + 22036055 UGGCAGCAAGAUGCUGCCCCACUUUGAUCUCUACUCCUGUUGGCGGGAGAUUAAAGUGGUUCCUCGAUACGAAAACGCCACUGGGAGCAACAGCUCGGC--------------------- .((((((.....))))))((((((((((((((...((....))..)))))))))))))).................(((.....((((....)))))))--------------------- ( -40.70, z-score = -3.47, R) >consensus UGGCAGCAAAAUGCUGCCCCACUUUGAUCUGUACUCCUGUUGGCGGGAGAUUAAAGUGGUUCCUCGAUACGAGAACGCCACUGGGAGCAACAGCUCGGC_____________________ .((((((.....)))))).....((((.((...((((((....))))))......(((((................)))))..........)).))))...................... (-18.51 = -18.01 + -0.49)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:43 2011