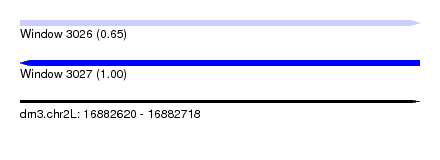
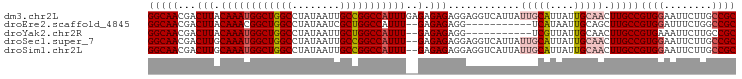
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,882,620 – 16,882,718 |
| Length | 98 |
| Max. P | 0.996234 |

| Location | 16,882,620 – 16,882,718 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 87.04 |
| Shannon entropy | 0.21237 |
| G+C content | 0.49410 |
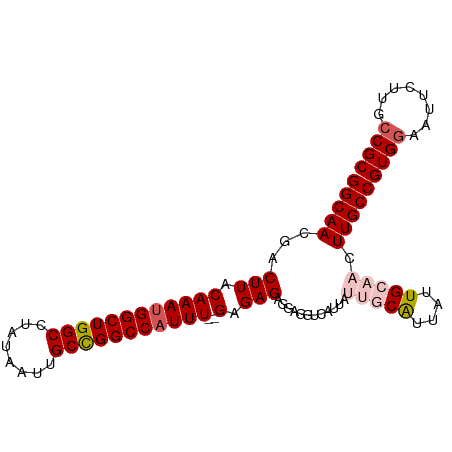
| Mean single sequence MFE | -28.30 |
| Consensus MFE | -20.38 |
| Energy contribution | -22.42 |
| Covariance contribution | 2.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.647333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16882620 98 + 23011544 GCGGCAAGAAUUCCACGGCAAGUUGCAAUAAUGCAAUAAUGACCUCCUCUCUCUCAAAUGGCCGGCAAUUAUAGGCCAGCCAUUUGUAAGUCGUUGCC ..(((((((.......((((.((((((....))))))..)).))......((..((((((((.(((........))).))))))))..)))).))))) ( -31.80, z-score = -2.92, R) >droEre2.scaffold_4845 15237698 85 - 22589142 GCGGCCAGAAAUCCACGGCAAGCUGCAAUUAUGA-----------CCUCUCUC--AAAUGGCCAGCGAUUAUAGGCCAGCCGUUUGUAAGUCGUUGCC (((((................)))))......((-----------(..((..(--(((((((..((........))..))))))))..))..)))... ( -23.09, z-score = -0.32, R) >droYak2.chr2R 16916703 85 - 21139217 GCGGCAAGAAUUUCACGGCAAGUUGCAAUAACGA-----------CCUCUCUC--AAAUGGCCAGCAAUUAUAGGCCAGCCAUUUGUAAGUCGUUGCC ..(((............((.....))...(((((-----------(......(--(((((((..((........))..))))))))...))))))))) ( -24.60, z-score = -1.94, R) >droSec1.super_7 519569 96 + 3727775 GCGGCAAGAAUUCCACGGCAAGUUGCAAUAAUGCAAUAAUGACCUCCUCUCUC--AAAUGGCCGGCAAUUAUAGGCCAGCCAUUUGCAAGUCGUUGCC ..(((((((.......((((.((((((....))))))..)).))....((..(--(((((((.(((........))).))))))))..)))).))))) ( -31.00, z-score = -2.34, R) >droSim1.chr2L 16566333 96 + 22036055 GCGGCAAGAAUUCCACGGCAAGUUGCAAUAAUGCAAUAAUGACCUCCUCUCUC--AAAUGGCCGGCAAUUAUAGGCCAGCCAUUUGCAAGUCGUUGCC ..(((((((.......((((.((((((....))))))..)).))....((..(--(((((((.(((........))).))))))))..)))).))))) ( -31.00, z-score = -2.34, R) >consensus GCGGCAAGAAUUCCACGGCAAGUUGCAAUAAUGCAAUAAUGACCUCCUCUCUC__AAAUGGCCGGCAAUUAUAGGCCAGCCAUUUGUAAGUCGUUGCC ..((((........(((((..(((((......)))))..................(((((((.(((........))).)))))))....))))))))) (-20.38 = -22.42 + 2.04)
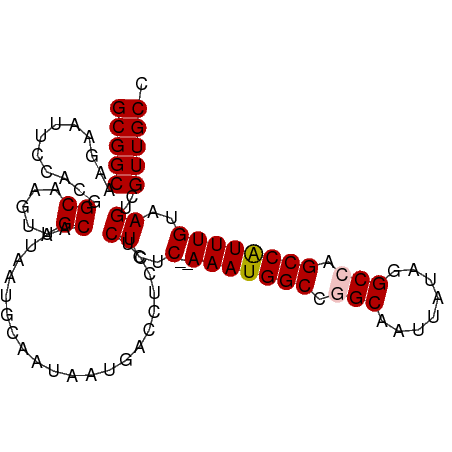
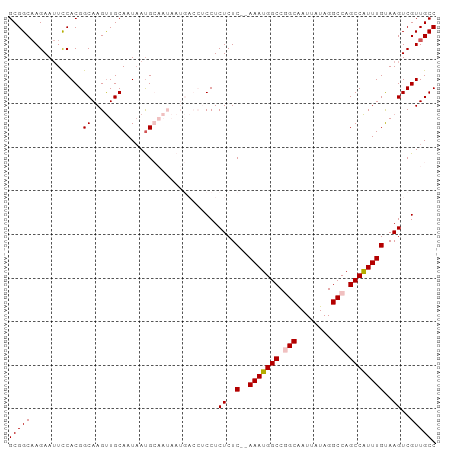
| Location | 16,882,620 – 16,882,718 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 87.04 |
| Shannon entropy | 0.21237 |
| G+C content | 0.49410 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -26.46 |
| Energy contribution | -27.66 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.90 |
| SVM RNA-class probability | 0.996234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16882620 98 - 23011544 GGCAACGACUUACAAAUGGCUGGCCUAUAAUUGCCGGCCAUUUGAGAGAGAGGAGGUCAUUAUUGCAUUAUUGCAACUUGCCGUGGAAUUCUUGCCGC (((((.(((((.((((((((((((........)))))))))))).)))......((.((...(((((....)))))..)))).......))))))).. ( -38.70, z-score = -4.18, R) >droEre2.scaffold_4845 15237698 85 + 22589142 GGCAACGACUUACAAACGGCUGGCCUAUAAUCGCUGGCCAUUU--GAGAGAGG-----------UCAUAAUUGCAGCUUGCCGUGGAUUUCUGGCCGC (((.........((((.(((..((........))..))).)))--)..(((((-----------((...((.((.....)).)).))))))).))).. ( -28.20, z-score = -1.33, R) >droYak2.chr2R 16916703 85 + 21139217 GGCAACGACUUACAAAUGGCUGGCCUAUAAUUGCUGGCCAUUU--GAGAGAGG-----------UCGUUAUUGCAACUUGCCGUGAAAUUCUUGCCGC (((((((((((.((((((((..((........))..)))))))--)....)))-----------)))((((.((.....)).)))).....))))).. ( -34.10, z-score = -4.41, R) >droSec1.super_7 519569 96 - 3727775 GGCAACGACUUGCAAAUGGCUGGCCUAUAAUUGCCGGCCAUUU--GAGAGAGGAGGUCAUUAUUGCAUUAUUGCAACUUGCCGUGGAAUUCUUGCCGC ((((((..(((.((((((((((((........)))))))))))--)...)))..).......(((((....))))).)))))((((........)))) ( -39.30, z-score = -3.93, R) >droSim1.chr2L 16566333 96 - 22036055 GGCAACGACUUGCAAAUGGCUGGCCUAUAAUUGCCGGCCAUUU--GAGAGAGGAGGUCAUUAUUGCAUUAUUGCAACUUGCCGUGGAAUUCUUGCCGC ((((((..(((.((((((((((((........)))))))))))--)...)))..).......(((((....))))).)))))((((........)))) ( -39.30, z-score = -3.93, R) >consensus GGCAACGACUUACAAAUGGCUGGCCUAUAAUUGCCGGCCAUUU__GAGAGAGGAGGUCAUUAUUGCAUUAUUGCAACUUGCCGUGGAAUUCUUGCCGC (((((...(((..(((((((((((........)))))))))))..)))..............(((((....))))).)))))((((........)))) (-26.46 = -27.66 + 1.20)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:40 2011