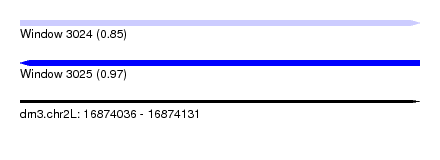
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,874,036 – 16,874,131 |
| Length | 95 |
| Max. P | 0.972299 |

| Location | 16,874,036 – 16,874,131 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 72.97 |
| Shannon entropy | 0.54799 |
| G+C content | 0.42133 |
| Mean single sequence MFE | -26.50 |
| Consensus MFE | -10.58 |
| Energy contribution | -11.49 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.847755 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16874036 95 + 23011544 --GUAUAACGAAAUAAUGAC--GUUUGAUUGGCCUUCGGCAAGGCCGUUUGCUGUUU---GUUGGCCAACUGUCAACGUCUUGUAAAA-UUUAUUGGCCAAUA --..................--.....(((((((..((((...))))(((((....(---((((((.....)))))))....))))).-......))))))). ( -25.40, z-score = -1.41, R) >droSim1.chr2L 16557528 91 + 22036055 --GUAAAGCGAAAUAAUGAC--GUUUGAUUGGCCUUCGGCAAGGCCGUUUGCUGUUGGCCAACU-------GUCAACGUCUUGUAAAA-UUUAUUGGCCAAUA --(((((.....((((.(((--(((.((((((((..(((((((....)))))))..)))))...-------)))))))))))))....-)))))......... ( -27.20, z-score = -1.98, R) >droSec1.super_7 511128 91 + 3727775 --GUAAAGCGAAAUAAUGAC--GUUUGAUUGGCCUUCGGCAAGGCCGUUUGCUGUUGGCCAACU-------GUCAACGUCUUGUAAAA-UUUAUUGGCCAAUA --(((((.....((((.(((--(((.((((((((..(((((((....)))))))..)))))...-------)))))))))))))....-)))))......... ( -27.20, z-score = -1.98, R) >droYak2.chr2R 16908208 90 - 21139217 ---GAAAACGAAAUAAUGAC--GUUUGAUUGGCCUUCGGCAAGGCCGUUUGCUGUUGGCCAACU-------GUCAACGUCUUGUAAAA-UUUAUUGGCCAAUA ---.........((((.(((--(((.((((((((..(((((((....)))))))..)))))...-------)))))))))))))....-.............. ( -26.80, z-score = -2.33, R) >droEre2.scaffold_4845 15229354 90 - 22589142 ---CCAAACGAAAUAAUGAC--GUUUGAUUGGCCUUCGGCAAGGCCGUUUGCUGUUGGCCAACU-------GUCAACGUCUUGUAAAA-UUUAUUGGCCAAUA ---.((((((.........)--)))))(((((((..((((...))))((((((((((((.....-------)))))))....))))).-......))))))). ( -27.50, z-score = -2.42, R) >droAna3.scaffold_12916 7433870 97 + 16180835 --AUAAAGCCAAAUAAUGAC--GUUUGAUUGGCCUUUGGCAAGGCCGUUUGAUGUUGGCCAACG-CCAAAUGUCAACGUCUUGUAAAA-UUUAUUGGCCAAUA --.....(((((((((.(((--((.....((((.((((((..(((((........)))))...)-))))).)))))))))))))....-....)))))..... ( -31.00, z-score = -2.75, R) >dp4.chr4_group3 8794150 92 - 11692001 --------CCAGAUAAUGAC--GUUUGAUUGGCCUUUGGCAAGGCCGUUUUAGCCUGUCGGUUGGCCAAAUGUCAACGUCUUGUAAAA-UUUAUUGGCCAAUA --------((((((((.(((--(((.((((((((.((((((.(((.......)))))))))..)))))...)))))))))))))....-....))))...... ( -28.50, z-score = -2.11, R) >droPer1.super_1 5900432 92 - 10282868 --------CCAGAUAAUGAC--GUUUGAUUGGCCUUUGGCAAGGCCGUUUUAGCCUGUCGGUUGGCCAAAUGUCAACGUCUUGUAAAA-UUUAUUGGCCAAUA --------((((((((.(((--(((.((((((((.((((((.(((.......)))))))))..)))))...)))))))))))))....-....))))...... ( -28.50, z-score = -2.11, R) >droWil1.scaffold_180764 1259546 85 + 3949147 --------UGACGUUUUGCCUGGUUUGAUUGGCCUUUGGCAAGGCCGCUUCAUUCCUGGGGUUG----------AAUGUCUUGUAAAAAUUUAUUGGCCAAUA --------.(((((((.((((((..(((.(((((((....)))))))..)))..))..)))).)----------))))))....................... ( -27.60, z-score = -2.18, R) >droVir3.scaffold_12963 7786965 93 - 20206255 --ACAUAUUUGUGUAAUGAC-GCUUUGAUUGGC-UCGACCUUGACCAAGAGAAUCCGUUGGCC-----AUCGUCAACGACUUGUAAAA-UUUAUUGGCCAAUA --........((((....))-))....((((((-.(((.(((......)))....(((((((.-----...)))))))..........-....))))))))). ( -20.10, z-score = -0.73, R) >droGri2.scaffold_15126 2491948 96 - 8399593 AUACAUAUUUAUGUAAUGAC-GCCUUGAUUGGC-UCGACCUUGACCAAGAAAAUC-GUUGGCCG---CAUCGUCAACGACUUGUAAAA-UUUAUUGGCCAAUA .(((((....))))).....-......((((((-(..........((((....((-((((((..---....)))))))))))).....-......))))))). ( -21.70, z-score = -1.42, R) >consensus ___UAAA_CGAAAUAAUGAC__GUUUGAUUGGCCUUCGGCAAGGCCGUUUGAUGUUGGCCGACG____A__GUCAACGUCUUGUAAAA_UUUAUUGGCCAAUA ...........................(((((((....(((((((................................)))))))...........))))))). (-10.58 = -11.49 + 0.91)

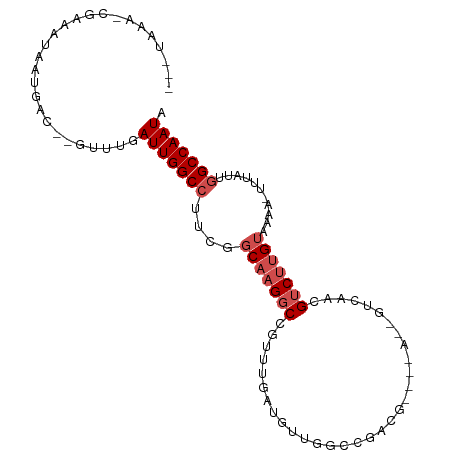
| Location | 16,874,036 – 16,874,131 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 72.97 |
| Shannon entropy | 0.54799 |
| G+C content | 0.42133 |
| Mean single sequence MFE | -24.29 |
| Consensus MFE | -10.20 |
| Energy contribution | -10.35 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16874036 95 - 23011544 UAUUGGCCAAUAAA-UUUUACAAGACGUUGACAGUUGGCCAAC---AAACAGCAAACGGCCUUGCCGAAGGCCAAUCAAAC--GUCAUUAUUUCGUUAUAC-- ..(((((((((...-....((.....)).....))))))))).---...........((((((....)))))).....(((--(.........))))....-- ( -22.60, z-score = -2.07, R) >droSim1.chr2L 16557528 91 - 22036055 UAUUGGCCAAUAAA-UUUUACAAGACGUUGAC-------AGUUGGCCAACAGCAAACGGCCUUGCCGAAGGCCAAUCAAAC--GUCAUUAUUUCGCUUUAC-- ..............-........((((((...-------.(((((((..(.((((......)))).)..)))))))..)))--)))...............-- ( -23.70, z-score = -2.42, R) >droSec1.super_7 511128 91 - 3727775 UAUUGGCCAAUAAA-UUUUACAAGACGUUGAC-------AGUUGGCCAACAGCAAACGGCCUUGCCGAAGGCCAAUCAAAC--GUCAUUAUUUCGCUUUAC-- ..............-........((((((...-------.(((((((..(.((((......)))).)..)))))))..)))--)))...............-- ( -23.70, z-score = -2.42, R) >droYak2.chr2R 16908208 90 + 21139217 UAUUGGCCAAUAAA-UUUUACAAGACGUUGAC-------AGUUGGCCAACAGCAAACGGCCUUGCCGAAGGCCAAUCAAAC--GUCAUUAUUUCGUUUUC--- ..............-........((((((...-------.(((((((..(.((((......)))).)..)))))))..)))--)))..............--- ( -23.70, z-score = -2.45, R) >droEre2.scaffold_4845 15229354 90 + 22589142 UAUUGGCCAAUAAA-UUUUACAAGACGUUGAC-------AGUUGGCCAACAGCAAACGGCCUUGCCGAAGGCCAAUCAAAC--GUCAUUAUUUCGUUUGG--- ..(((((((((...-....((.....))....-------.)))))))))........((((((....))))))...(((((--(.........)))))).--- ( -26.50, z-score = -2.83, R) >droAna3.scaffold_12916 7433870 97 - 16180835 UAUUGGCCAAUAAA-UUUUACAAGACGUUGACAUUUGG-CGUUGGCCAACAUCAAACGGCCUUGCCAAAGGCCAAUCAAAC--GUCAUUAUUUGGCUUUAU-- .(((((((......-.....(((....)))...(((((-((..((((..........)))).)))))))))))))).....--((((.....)))).....-- ( -29.10, z-score = -3.08, R) >dp4.chr4_group3 8794150 92 + 11692001 UAUUGGCCAAUAAA-UUUUACAAGACGUUGACAUUUGGCCAACCGACAGGCUAAAACGGCCUUGCCAAAGGCCAAUCAAAC--GUCAUUAUCUGG-------- ..............-........((((((((..(((((((........)))))))..((((((....))))))..)).)))--))).........-------- ( -27.30, z-score = -3.29, R) >droPer1.super_1 5900432 92 + 10282868 UAUUGGCCAAUAAA-UUUUACAAGACGUUGACAUUUGGCCAACCGACAGGCUAAAACGGCCUUGCCAAAGGCCAAUCAAAC--GUCAUUAUCUGG-------- ..............-........((((((((..(((((((........)))))))..((((((....))))))..)).)))--))).........-------- ( -27.30, z-score = -3.29, R) >droWil1.scaffold_180764 1259546 85 - 3949147 UAUUGGCCAAUAAAUUUUUACAAGACAUU----------CAACCCCAGGAAUGAAGCGGCCUUGCCAAAGGCCAAUCAAACCAGGCAAAACGUCA-------- .....(((.................((((----------(........)))))....((((((....))))))..........))).........-------- ( -20.40, z-score = -1.85, R) >droVir3.scaffold_12963 7786965 93 + 20206255 UAUUGGCCAAUAAA-UUUUACAAGUCGUUGACGAU-----GGCCAACGGAUUCUCUUGGUCAAGGUCGA-GCCAAUCAAAGC-GUCAUUACACAAAUAUGU-- ..((((((......-........((((....))))-----((((((.((....))))))))..))))))-((........))-..................-- ( -18.30, z-score = -0.24, R) >droGri2.scaffold_15126 2491948 96 + 8399593 UAUUGGCCAAUAAA-UUUUACAAGUCGUUGACGAUG---CGGCCAAC-GAUUUUCUUGGUCAAGGUCGA-GCCAAUCAAGGC-GUCAUUACAUAAAUAUGUAU ..((((((......-........((((....)))).---.((((((.-(.....)))))))..))))))-(((......)))-.....(((((....))))). ( -24.60, z-score = -1.95, R) >consensus UAUUGGCCAAUAAA_UUUUACAAGACGUUGAC__U____CGUCCGCCAACAUCAAACGGCCUUGCCGAAGGCCAAUCAAAC__GUCAUUAUUUCG_UUUA___ .(((((((............(((....)))...........................(((...)))...)))))))........................... (-10.20 = -10.35 + 0.16)
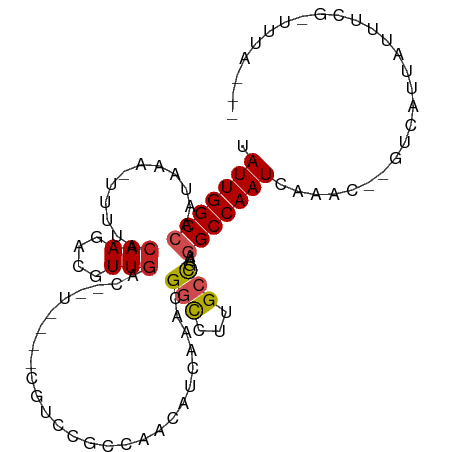
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:38 2011