| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,808,658 – 16,808,760 |
| Length | 102 |
| Max. P | 0.530514 |

| Location | 16,808,658 – 16,808,760 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 72.62 |
| Shannon entropy | 0.55288 |
| G+C content | 0.41208 |
| Mean single sequence MFE | -25.10 |
| Consensus MFE | -10.33 |
| Energy contribution | -10.76 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.530514 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
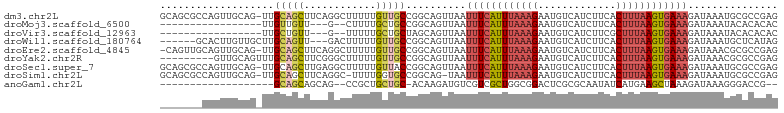
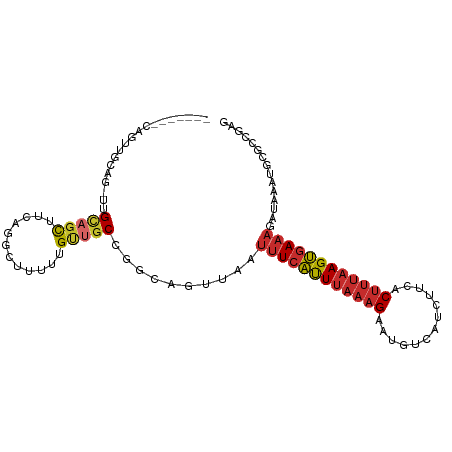
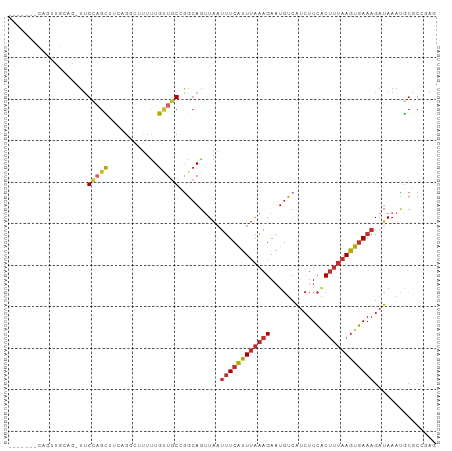
>dm3.chr2L 16808658 102 - 23011544 GCAGCGCCAGUUGCAG-UUGCAGCUUCAGGCUUUUUGUUGCCGGCAGUUAAUUUCAUUUAAAGAAUGUCAUCUUCACUUUAAGUGAAAGAUAAAUGCGCCGAG ((((((((((((((..-..))))))...))).....)))))((((.(((..((((((((((((..((.......))))))))))))))....)))..)))).. ( -32.20, z-score = -2.52, R) >droMoj3.scaffold_6500 26635677 81 + 32352404 -----------------UUGUUGUU---G--CUUUUGCUGCCGGCAGUUAAUUUCAUUUAAAGAAUGUCAUCUUCACUUUAAGUGAAAGAUAAAUACACACAC -----------------((((((..---(--(....))...))))))....((((((((((((..((.......))))))))))))))............... ( -14.80, z-score = -0.67, R) >droVir3.scaffold_12963 13926897 81 - 20206255 -----------------UUGCUGUU---G--UUUUUGCUGCUAGCAGUUAAUUUCAUUUAAAGAAUGUCAUCUUCGCUUUAAGUGAAAGAUAAAUACACACAC -----------------(((((((.---(--(....)).)).)))))....((((((((((((...(....)....))))))))))))............... ( -15.70, z-score = -0.73, R) >droWil1.scaffold_180764 1176117 94 - 3949147 ------GCACUUGUUGCUUGCAGUU---GACUUUUUGUUGCCGGCAGUUAAUUUCAUUUAAAGAAUGUCAUCUUCACUUUAAGUGAAAGAUAAAUGCUCAUAG ------(((.((((((((.((((..---.........)))).)))......((((((((((((..((.......))))))))))))))))))).)))...... ( -20.90, z-score = -0.97, R) >droEre2.scaffold_4845 19879680 101 - 22589142 -CAGUUGCAGUUGCAG-UUGCAGCUUCAGGCUUUUUGUUGCCGGCAGUUAAUUUCAUUUAAAGAAUGUCAUCUUCACUUUAAGUGAAAGAUAAACGCGCCGAG -.((((((((......-))))))))((.(((........)))(((.(((..((((((((((((..((.......))))))))))))))....)))..))))). ( -30.30, z-score = -2.47, R) >droYak2.chr2R 16846731 94 + 21139217 ---------GUUGCAGUUUGCAGCUUCGGGCUUUUUGUUGCCGGCAGUUAAUUUCAUUUAAAGAAUGUCAUCUUCACUUUAAGUGAAAGAUAAACGCGCCGAG ---------(((((.....))))).((.(((........)))(((.(((..((((((((((((..((.......))))))))))))))....)))..))))). ( -27.80, z-score = -2.33, R) >droSec1.super_7 454762 102 - 3727775 GCAGCGCCAGUUGCAG-UUGCAGCUUGAGGCUUUUUGUUACCGGCAGUUAAUUUCAUUUAAAGAAUGUCAUCUUCACUUUAAGUGAAAGAUAAAUGCGCCGAG .....(((((((((..-..))))))...)))..........((((.(((..((((((((((((..((.......))))))))))))))....)))..)))).. ( -28.30, z-score = -1.47, R) >droSim1.chr2L 16501099 100 - 22036055 GCAGCGCCAGUUGCAG-UUGCAGCUUCAGGC-UUUUGGUGCCGGCAG-UAAUUUCAUUUAAAGAAUGUCAUCUUCACUUUAAGUGAAAGAUAAAUGCGCCGAG .....(((((((((..-..))))))...)))-..(((((((......-...((((((((((((..((.......)))))))))))))).......))))))). ( -30.39, z-score = -1.72, R) >anoGam1.chr2L 37567526 79 - 48795086 -------------------GCAGCAGCAG--CCGCUGCUGC-ACAAGAUGUCGUCGCUGGCGGACUCGCGCAAUAUCAUGAAGCUAAAGAUAAAGGGACCG-- -------------------((((((((..--..))))))))-.((.(((((.(.(((.((....)).)))).))))).)).....................-- ( -25.50, z-score = -0.38, R) >consensus _______CAGUUGCAG_UUGCAGCUUCAGGCUUUUUGUUGCCGGCAGUUAAUUUCAUUUAAAGAAUGUCAUCUUCACUUUAAGUGAAAGAUAAAUGCGCCGAG ...................(((((............)))))..........((((((((((((.............))))))))))))............... (-10.33 = -10.76 + 0.44)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:33 2011