| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,782,846 – 16,782,983 |
| Length | 137 |
| Max. P | 0.880119 |

| Location | 16,782,846 – 16,782,966 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.38 |
| Shannon entropy | 0.18094 |
| G+C content | 0.54702 |
| Mean single sequence MFE | -39.66 |
| Consensus MFE | -30.34 |
| Energy contribution | -30.66 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.31 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.880119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16782846 120 + 23011544 CAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC ..((.((((((((.(((.....))).))(((((..((((((........)))))).((((...........)))).....(((((....)))))....))))).....))))))..)).. ( -41.70, z-score = -2.60, R) >droSim1.chrU 13345234 120 + 15797150 CAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC ..((.((((((((.(((.....))).))(((((..((((((........)))))).((((...........)))).....(((((....)))))....))))).....))))))..)).. ( -41.70, z-score = -2.60, R) >droSec1.super_7 429569 120 + 3727775 CAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC ..((.((((((((.(((.....))).))(((((..((((((........)))))).((((...........)))).....(((((....)))))....))))).....))))))..)).. ( -41.70, z-score = -2.60, R) >droEre2.scaffold_4845 19853430 120 + 22589142 CAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC ..((.((((((((.(((.....))).))(((((..((((((........)))))).((((...........)))).....(((((....)))))....))))).....))))))..)).. ( -41.70, z-score = -2.60, R) >droYak2.chr2R 16819947 120 - 21139217 CAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC ..((.((((((((.(((.....))).))(((((..((((((........)))))).((((...........)))).....(((((....)))))....))))).....))))))..)).. ( -41.70, z-score = -2.60, R) >droAna3.scaffold_12916 7342548 120 + 16180835 CAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAGCUGAAUGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC ..((.((((((((.(((.....))).))(((((..((((((.(.....))))))).((((...........)))).....(((((....)))))....))))).....))))))..)).. ( -41.80, z-score = -2.08, R) >droPer1.super_1 5798840 120 - 10282868 CAGGAAAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAGCUGAACAUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC ..((..(((((((.(((.....))).))(((((..((((((........)))))).((((...........)))).....(((((....)))))....))))).....)))))..))... ( -39.50, z-score = -2.10, R) >dp4.chr4_group3 8694511 120 - 11692001 CAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAGCUGAACAUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC ..((.((((((((.(((.....))).))(((((..((((((........)))))).((((...........)))).....(((((....)))))....))))).....))))))..)).. ( -42.50, z-score = -2.72, R) >droWil1.scaffold_180764 1140115 120 + 3949147 CAGGAGAUCUCUGGCUUGAAGAAGGAUAUCGAAGAUCUGGAAUUGAACAUCCAGAAGGCCGAGCAAGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC ..((.(((((((.(.((.(((..((..........((((((........)))))).((((...........)))).))..(((((....)))))....))).))).).))))))..)).. ( -38.30, z-score = -2.90, R) >droMoj3.scaffold_6500 2811019 120 + 32352404 CAGGAGAUCUCCGGCUUGAAGAAGGACAUCGAGGAUCUGGAAUUGAGCGUCCAGAAGGCCGAGCAGGAUAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC ..((.((((((((.(((.....)))...(((((..((((((........)))))).((((...........)))).....(((((....)))))....))))).))..))))))..)).. ( -38.30, z-score = -1.53, R) >droVir3.scaffold_12963 10130661 120 + 20206255 CAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAAUUGAGCGUCCAGAAGGCCGAACAGGAUAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC ..((.((((((((.(((.....))).))(((((..((((((........)))))).((((...........)))).....(((((....)))))....))))).....))))))..)).. ( -41.70, z-score = -2.64, R) >droGri2.scaffold_15252 13265152 120 + 17193109 CAAGAGAUCUCUGGCCUGAAGAAGGACAUCGAAGAUAUGGAGCUGAACAUCCAGAAGGCCGAACAGGAUAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC .....((((((((.(((.....))).))(((......(((..(((......)))..((((...........)))).))).(((((....)))))..........))).))))))...... ( -33.50, z-score = -1.92, R) >anoGam1.chr3R 50164750 120 + 53272125 CAGGAAAUCGGCAGCCAGAAGAAGGAUGCUGAGGACCUGGAACUGCAGAUCCAGAAGAUUGAGCAGGACAAGGCCUCGAAGGAUCACCAGAUCCGCAACUUGAAUGAUGAGAUCGCCCAC ..((..(((..((..((.....(((.((((((((.(((.(..((((.((((.....))))..))))..).))))))))..(((((....)))))))).)))...)).)).)))..))... ( -38.40, z-score = -2.18, R) >apiMel3.GroupUn 689516 120 + 399230636 CAGGAAGUGGCGGGCCUGAAGAAAGAUAUCGAGGAUUUGGAACUUAACCUGCAAAAGUCUGAGCAGGACAAGGCGACCAAGGAUCAUCAGAUCCGUAACUUGAACGACGAGAUCGCUCAU ....((((.((((..((((.((......))((...(((((..(((..(((((..........)))))..)))....)))))..)).))))..)))).))))((.(((.....))).)).. ( -32.70, z-score = -0.94, R) >consensus CAGGAGAUCUCUGGCCUGAAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC ..((..(((((((.(((.....))).))(((((..((((((........)))))).((((...........)))).....(((((....)))))....))))).....)))))..))... (-30.34 = -30.66 + 0.33)

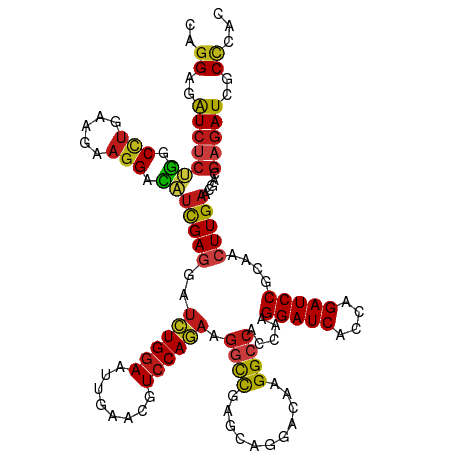

| Location | 16,782,846 – 16,782,966 |
|---|---|
| Length | 120 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.38 |
| Shannon entropy | 0.18094 |
| G+C content | 0.54702 |
| Mean single sequence MFE | -41.20 |
| Consensus MFE | -34.66 |
| Energy contribution | -34.94 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614082 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16782846 120 - 23011544 GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUG ..(((.((((((.........((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))(.(((.....))).).)))))).))). ( -43.60, z-score = -1.85, R) >droSim1.chrU 13345234 120 - 15797150 GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUG ..(((.((((((.........((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))(.(((.....))).).)))))).))). ( -43.60, z-score = -1.85, R) >droSec1.super_7 429569 120 - 3727775 GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUG ..(((.((((((.........((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))(.(((.....))).).)))))).))). ( -43.60, z-score = -1.85, R) >droEre2.scaffold_4845 19853430 120 - 22589142 GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUG ..(((.((((((.........((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))(.(((.....))).).)))))).))). ( -43.60, z-score = -1.85, R) >droYak2.chr2R 16819947 120 + 21139217 GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUG ..(((.((((((.........((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))(.(((.....))).).)))))).))). ( -43.60, z-score = -1.85, R) >droAna3.scaffold_12916 7342548 120 - 16180835 GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACAUUCAGCUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUG ..(((.((((((((.(..(((((..((((((....)))))...)..).))))..).))..(((((...(((((((.((.........)).)))))))......))))).)))))).))). ( -44.50, z-score = -1.55, R) >droPer1.super_1 5798840 120 + 10282868 GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGAUGUUCAGCUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUUUCCUG ..(((.((((((((.(..(((((..((((((....)))))...)..).))))..).))..(((((((((((.(.....))))))).....((......))...))))).)))))).))). ( -41.50, z-score = -0.93, R) >dp4.chr4_group3 8694511 120 + 11692001 GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGAUGUUCAGCUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUG ..(((.((((((((.(..(((((..((((((....)))))...)..).))))..).))..(((((((((((.(.....))))))).....((......))...))))).)))))).))). ( -43.70, z-score = -1.37, R) >droWil1.scaffold_180764 1140115 120 - 3949147 GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCUUGCUCGGCCUUCUGGAUGUUCAAUUCCAGAUCUUCGAUAUCCUUCUUCAAGCCAGAGAUCUCCUG ..(((.((((((..............(((((....))))).((((((((..(.....)..)))).((((((........))))))....................)))))))))).))). ( -40.30, z-score = -1.94, R) >droMoj3.scaffold_6500 2811019 120 - 32352404 GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGCUCGGCCUUCUGGACGCUCAAUUCCAGAUCCUCGAUGUCCUUCUUCAAGCCGGAGAUCUCCUG ..(((.((((((............((((((..(((.((.....)).)))..)))).)).((((..((((((........)))))).....((......)).....)))))))))).))). ( -39.80, z-score = -0.96, R) >droVir3.scaffold_12963 10130661 120 - 20206255 GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGUUCGGCCUUCUGGACGCUCAAUUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUG ..(((.((((((.........((((.((((((((((((((..((.((((...........)))).)).)))...)))...)))))))).))))(.(((.....))).).)))))).))). ( -42.30, z-score = -1.65, R) >droGri2.scaffold_15252 13265152 120 - 17193109 GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGUUCGGCCUUCUGGAUGUUCAGCUCCAUAUCUUCGAUGUCCUUCUUCAGGCCAGAGAUCUCUUG ..(((.((((((((((...(((.((.(((.((((..((((..((.((((...........)))).)).))))..)))).))).)).)))))))(.(((.....))).).)))))).))). ( -35.90, z-score = -0.35, R) >anoGam1.chr3R 50164750 120 - 53272125 GUGGGCGAUCUCAUCAUUCAAGUUGCGGAUCUGGUGAUCCUUCGAGGCCUUGUCCUGCUCAAUCUUCUGGAUCUGCAGUUCCAGGUCCUCAGCAUCCUUCUUCUGGCUGCCGAUUUCCUG ..(((.((((................(((((....)))))...(((((((.(..((((...(((.....)))..))))..).))).)))).(((.((.......)).))).)))).))). ( -37.30, z-score = -1.04, R) >apiMel3.GroupUn 689516 120 - 399230636 AUGAGCGAUCUCGUCGUUCAAGUUACGGAUCUGAUGAUCCUUGGUCGCCUUGUCCUGCUCAGACUUUUGCAGGUUAAGUUCCAAAUCCUCGAUAUCUUUCUUCAGGCCCGCCACUUCCUG .((((((((...))))))))(((..(((..((((.(((..((((....((((.(((((..........))))).))))..)))))))...((......)).))))..)))..)))..... ( -33.50, z-score = -2.13, R) >consensus GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUUCAGGCCAGAGAUCUCCUG ..(((.((((((............((((((..(((.((.....)).)))..)))).))..(((((((((((........)))))).....((......))...))))).)))))).))). (-34.66 = -34.94 + 0.28)

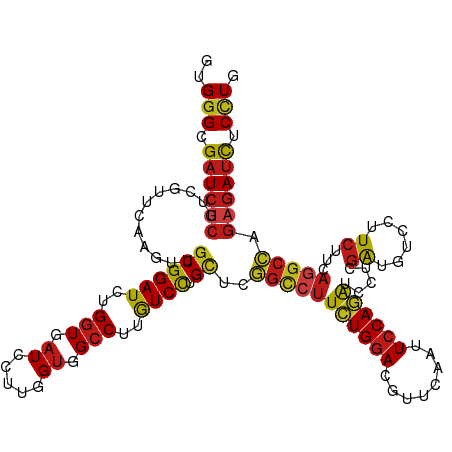

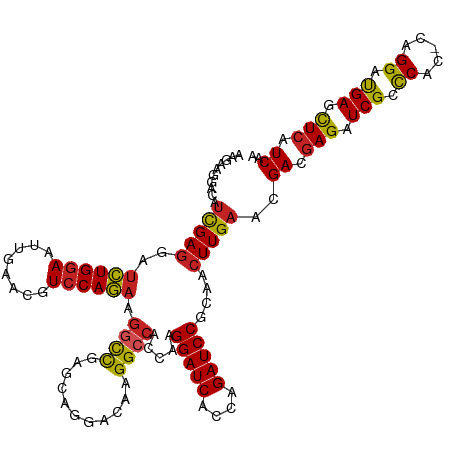
| Location | 16,782,864 – 16,782,983 |
|---|---|
| Length | 119 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.20 |
| Shannon entropy | 0.15730 |
| G+C content | 0.52442 |
| Mean single sequence MFE | -35.70 |
| Consensus MFE | -30.69 |
| Energy contribution | -30.69 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857225 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16782864 119 + 23011544 AAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC-CAGGAUGAGCUCAUCAA ..........(((((..((((((........)))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((..-..)).))).))).)).. ( -35.30, z-score = -1.69, R) >droSim1.chrU 13345252 120 + 15797150 AAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCACACAGGAUGAGCUCAUCAA ..........(((((..((((((........)))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((.....)).))).))).)).. ( -35.30, z-score = -1.92, R) >droSec1.super_7 429587 119 + 3727775 AAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC-CAGGAUGAGCUCAUCAA ..........(((((..((((((........)))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((..-..)).))).))).)).. ( -35.30, z-score = -1.69, R) >droEre2.scaffold_4845 19853448 119 + 22589142 AAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC-CAGGAUGAGCUCAUCAA ..........(((((..((((((........)))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((..-..)).))).))).)).. ( -35.30, z-score = -1.69, R) >droYak2.chr2R 16819965 119 - 21139217 AAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC-CAGGAUGAGCUCAUCAA ..........(((((..((((((........)))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((..-..)).))).))).)).. ( -35.30, z-score = -1.69, R) >droAna3.scaffold_12916 7342566 119 + 16180835 AAGAAGGACAUCGAGGAUCUGGAGCUGAAUGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC-CAGGAUGAGCUCAUCAA ..........(((((..((((((.(.....))))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((..-..)).))).))).)).. ( -35.40, z-score = -1.14, R) >droWil1.scaffold_180764 1140133 119 + 3949147 AAGAAGGAUAUCGAAGAUCUGGAAUUGAACAUCCAGAAGGCCGAGCAAGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC-CAGGAUGAGCUCAUCAA (((..((..........((((((........)))))).((((...........)))).))..(((((....)))))....)))....((.(((.(((.((..-..)).))).))).)).. ( -33.10, z-score = -2.32, R) >droMoj3.scaffold_6500 2811037 119 + 32352404 AAGAAGGACAUCGAGGAUCUGGAAUUGAGCGUCCAGAAGGCCGAGCAGGAUAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC-CAGGAUGAGCUCAUCAA ..........(((((..((((((........)))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((..-..)).))).))).)).. ( -35.30, z-score = -1.41, R) >droVir3.scaffold_12963 10130679 119 + 20206255 AAGAAGGACAUCGAGGAUCUGGAAUUGAGCGUCCAGAAGGCCGAACAGGAUAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC-CAGGAUGAGCUCAUCAA ..........(((((..((((((........)))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((..-..)).))).))).)).. ( -35.30, z-score = -1.74, R) >anoGam1.chr3R 50164768 119 + 53272125 AAGAAGGAUGCUGAGGACCUGGAACUGCAGAUCCAGAAGAUUGAGCAGGACAAGGCCUCGAAGGAUCACCAGAUCCGCAACUUGAAUGAUGAGAUCGCCCAC-CAGGAUGAGCUCAUCAA ....(((.((((((((.(((.(..((((.((((.....))))..))))..).))))))))..(((((....)))))))).)))...(((((((.(((.((..-..)).))).))))))). ( -45.10, z-score = -3.86, R) >apiMel3.GroupUn 689534 119 + 399230636 AAGAAAGAUAUCGAGGAUUUGGAACUUAACCUGCAAAAGUCUGAGCAGGACAAGGCGACCAAGGAUCAUCAGAUCCGUAACUUGAACGACGAGAUCGCUCAU-CAGGACGAGUUGAUCAA ..........(((((...((((..(((..(((((..........)))))..)))....))))(((((....)))))....))))).......((((((((..-......))).))))).. ( -32.00, z-score = -1.49, R) >consensus AAGAAGGACAUCGAGGAUCUGGAAUUGAACGUCCAGAAGGCCGAGCAGGACAAGGCCACCAAGGAUCACCAGAUCCGCAACUUGAACGACGAGAUCGCCCAC_CAGGAUGAGCUCAUCAA ..........(((((..((((((........)))))).((((...........)))).....(((((....)))))....)))))..((.(((.(((.((.....)).))).))).)).. (-30.69 = -30.69 + 0.01)
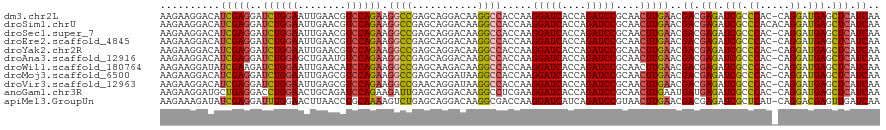

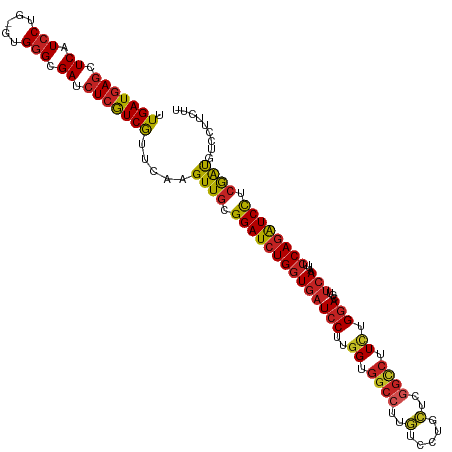
| Location | 16,782,864 – 16,782,983 |
|---|---|
| Length | 119 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.20 |
| Shannon entropy | 0.15730 |
| G+C content | 0.52442 |
| Mean single sequence MFE | -40.70 |
| Consensus MFE | -34.40 |
| Energy contribution | -34.98 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.795882 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16782864 119 - 23011544 UUGAUGAGCUCAUCCUG-GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUU ..((((((.((..((..-..))..)).))))))......((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))......... ( -41.90, z-score = -1.86, R) >droSim1.chrU 13345252 120 - 15797150 UUGAUGAGCUCAUCCUGUGUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUU ..((((((.((..((.....))..)).))))))......((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))......... ( -41.90, z-score = -2.21, R) >droSec1.super_7 429587 119 - 3727775 UUGAUGAGCUCAUCCUG-GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUU ..((((((.((..((..-..))..)).))))))......((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))......... ( -41.90, z-score = -1.86, R) >droEre2.scaffold_4845 19853448 119 - 22589142 UUGAUGAGCUCAUCCUG-GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUU ..((((((.((..((..-..))..)).))))))......((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))......... ( -41.90, z-score = -1.86, R) >droYak2.chr2R 16819965 119 + 21139217 UUGAUGAGCUCAUCCUG-GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUU ..((((((.((..((..-..))..)).))))))......((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))......... ( -41.90, z-score = -1.86, R) >droAna3.scaffold_12916 7342566 119 - 16180835 UUGAUGAGCUCAUCCUG-GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACAUUCAGCUCCAGAUCCUCGAUGUCCUUCUU ..((((((.((..((..-..))..)).))))))......((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))......... ( -41.90, z-score = -1.31, R) >droWil1.scaffold_180764 1140133 119 - 3949147 UUGAUGAGCUCAUCCUG-GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCUUGCUCGGCCUUCUGGAUGUUCAAUUCCAGAUCUUCGAUAUCCUUCUU ..((((((.((..((..-..))..)).))))))......((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))......... ( -39.60, z-score = -1.79, R) >droMoj3.scaffold_6500 2811037 119 - 32352404 UUGAUGAGCUCAUCCUG-GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGCUCGGCCUUCUGGACGCUCAAUUCCAGAUCCUCGAUGUCCUUCUU ..((((((.((..((..-..))..)).))))))......((((.((((((((((((((..((.((((...........)))).)).)))...)))...)))))))).))))......... ( -40.60, z-score = -1.45, R) >droVir3.scaffold_12963 10130679 119 - 20206255 UUGAUGAGCUCAUCCUG-GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUAUCCUGUUCGGCCUUCUGGACGCUCAAUUCCAGAUCCUCGAUGUCCUUCUU ..((((((.((..((..-..))..)).))))))......((((.((((((((((((((..((.((((...........)))).)).)))...)))...)))))))).))))......... ( -40.60, z-score = -1.61, R) >anoGam1.chr3R 50164768 119 - 53272125 UUGAUGAGCUCAUCCUG-GUGGGCGAUCUCAUCAUUCAAGUUGCGGAUCUGGUGAUCCUUCGAGGCCUUGUCCUGCUCAAUCUUCUGGAUCUGCAGUUCCAGGUCCUCAGCAUCCUUCUU .(((((((.((..((..-..))..)).)))))))...(((.((((((((....)))))...(((((((.(..((((...(((.....)))..))))..).))).)))).)))..)))... ( -43.80, z-score = -2.41, R) >apiMel3.GroupUn 689534 119 - 399230636 UUGAUCAACUCGUCCUG-AUGAGCGAUCUCGUCGUUCAAGUUACGGAUCUGAUGAUCCUUGGUCGCCUUGUCCUGCUCAGACUUUUGCAGGUUAAGUUCCAAAUCCUCGAUAUCUUUCUU ..(((((.(..((((((-(((((((((...))))))))..))).))))..).))))).((((....((((.(((((..........))))).))))..)))).................. ( -31.70, z-score = -1.62, R) >consensus UUGAUGAGCUCAUCCUG_GUGGGCGAUCUCGUCGUUCAAGUUGCGGAUCUGGUGAUCCUUGGUGGCCUUGUCCUGCUCGGCCUUCUGGACGUUCAAUUCCAGAUCCUCGAUGUCCUUCUU .(((((((.((.(((.....))).)).))))))).....((((.((((((((((((((..((.((((..(.....)..)))).)).)))...)))...)))))))).))))......... (-34.40 = -34.98 + 0.59)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:31 2011