
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,780,348 – 16,780,478 |
| Length | 130 |
| Max. P | 0.999958 |

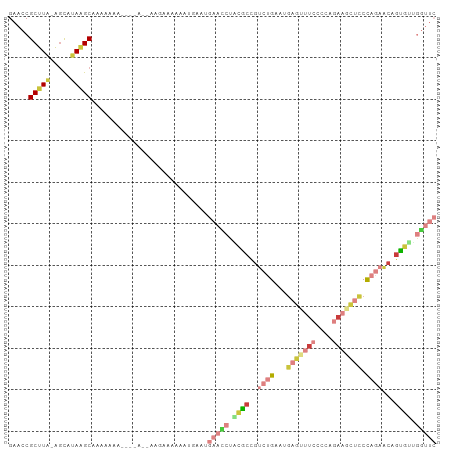
| Location | 16,780,348 – 16,780,449 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 67.29 |
| Shannon entropy | 0.64208 |
| G+C content | 0.41948 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -12.23 |
| Energy contribution | -15.26 |
| Covariance contribution | 3.03 |
| Combinations/Pair | 1.52 |
| Mean z-score | -4.36 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 5.24 |
| SVM RNA-class probability | 0.999958 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
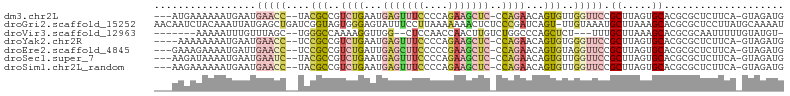
>dm3.chr2L 16780348 101 + 23011544 GAACCGCUUA-AGCAUAAGCAAAAAAA-GAAAAAAUGAAAAAAUGAAUGAACCUACGCCGUCUGAAUGAGUUUCCCCAGAAGCUCCCAGAACAGUGUUGGUUC .....(((((-....))))).......-....................(((((.((((..((((...(((((((....))))))).))))...)))).))))) ( -26.50, z-score = -5.05, R) >droGri2.scaffold_15252 13262383 96 + 17193109 GAAUCGCUUUCGCUUUAAGCAUAAAAA--CAAUCUACAAAUUAUGAGCUGAUCGGUAGUGGGAGUAUUUCCUUAAAAAAACUCUCCC-GAUCAGUUUGU---- .....((((.......)))).......--.............(..((((((((((....((((((.(((......))).))))))))-))))))))..)---- ( -26.00, z-score = -3.69, R) >droVir3.scaffold_12963 10128124 86 + 20206255 GAAUCGCUUGUAGCUUAAGCAAAAAAA-------------AAAUUUGUUUAGCUGGGCCAAAAGGUUG-GCUCCAACCAACUUGUCUGGCCCAGCUCUUU--- .....(((((.....))))).......-------------..........((((((((((..((((((-(......)))))))...))))))))))....--- ( -34.10, z-score = -4.69, R) >droYak2.chr2R 16817456 91 - 21139217 GAACCGCUUA-AGCAUAGGCCAAAAAA-----------AAAAAUGAAUGAACCUCCGCCGUCUGAAUGAGUUUCCCCAGAAGCUCCCAGAACAGUGUGGGUUC .....(((((-....))))).......-----------..........(((((..(((..((((...(((((((....))))))).))))...)))..))))) ( -25.80, z-score = -3.52, R) >droEre2.scaffold_4845 19851016 95 + 22589142 GAACCGCUUA-AGCAUAAGCAAAAAAA-------GAAAGAAAAUGAUUGAACCUCCGCCGUCUGAUUGAGCUUCCCCCGAAGCUCCCAGAACAGUGUAGGUUC .....(((((-....))))).......-------..............((((((.(((..((((...(((((((....))))))).))))...))).)))))) ( -28.70, z-score = -5.27, R) >droSec1.super_7 427082 98 + 3727775 GAACCGCUUA-AGCAUAAGCAAAAAAA----AAAAAGAUAAAAUGAAUGAAUCUACGCCGUCUGAAUGAGUUUCCCCAGAAGCUCCCAGAACAGUGUUGGUUC ((((((((((-....))))).......----.......................((((..((((...(((((((....))))))).))))...)))).))))) ( -24.50, z-score = -4.35, R) >droSim1.chr2L_random 622706 102 + 909653 GAACCGCUUA-AGCAUAUGCAAAAAAAAGAAAAAAAGAAAAAAUGAAUGAACCUACGCCGUCUGAAUGAGUUUCCCCAGAAGCUCCCAGAACAGUGUUGGUUC .....((...-.))..................................(((((.((((..((((...(((((((....))))))).))))...)))).))))) ( -23.60, z-score = -3.97, R) >consensus GAACCGCUUA_AGCAUAAGCAAAAAAA____A__AAGAAAAAAUGAAUGAACCUACGCCGUCUGAAUGAGUUUCCCCAGAAGCUCCCAGAACAGUGUUGGUUC .....(((((.....)))))............................(((((.((((..((((...(((((((....))))))).))))...)))).))))) (-12.23 = -15.26 + 3.03)


| Location | 16,780,348 – 16,780,449 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 67.29 |
| Shannon entropy | 0.64208 |
| G+C content | 0.41948 |
| Mean single sequence MFE | -27.70 |
| Consensus MFE | -7.99 |
| Energy contribution | -9.63 |
| Covariance contribution | 1.64 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.12 |
| SVM RNA-class probability | 0.982988 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16780348 101 - 23011544 GAACCAACACUGUUCUGGGAGCUUCUGGGGAAACUCAUUCAGACGGCGUAGGUUCAUUCAUUUUUUCAUUUUUUC-UUUUUUUGCUUAUGCU-UAAGCGGUUC (((((.((.((((.(((((......((((....))))))))))))).)).)))))....................-.....(((((((....-)))))))... ( -28.00, z-score = -3.40, R) >droGri2.scaffold_15252 13262383 96 - 17193109 ----ACAAACUGAUC-GGGAGAGUUUUUUUAAGGAAAUACUCCCACUACCGAUCAGCUCAUAAUUUGUAGAUUG--UUUUUAUGCUUAAAGCGAAAGCGAUUC ----(((((((((((-((..((((.((((....)))).))))......)))))))).......))))).(((((--(((((..((.....)))))))))))). ( -24.51, z-score = -2.70, R) >droVir3.scaffold_12963 10128124 86 - 20206255 ---AAAGAGCUGGGCCAGACAAGUUGGUUGGAGC-CAACCUUUUGGCCCAGCUAAACAAAUUU-------------UUUUUUUGCUUAAGCUACAAGCGAUUC ---....((((((((((((...((((((....))-))))..))))))))))))..........-------------.....((((((.......))))))... ( -34.40, z-score = -4.33, R) >droYak2.chr2R 16817456 91 + 21139217 GAACCCACACUGUUCUGGGAGCUUCUGGGGAAACUCAUUCAGACGGCGGAGGUUCAUUCAUUUUU-----------UUUUUUGGCCUAUGCU-UAAGCGGUUC (((((..(.((((.(((((......((((....))))))))))))).)..)))))..........-----------......((((...((.-...)))))). ( -24.60, z-score = -0.95, R) >droEre2.scaffold_4845 19851016 95 - 22589142 GAACCUACACUGUUCUGGGAGCUUCGGGGGAAGCUCAAUCAGACGGCGGAGGUUCAAUCAUUUUCUUUC-------UUUUUUUGCUUAUGCU-UAAGCGGUUC ((((((.(.((((.(((.(((((((....)))))))...))))))).).))))))..............-------.....(((((((....-)))))))... ( -33.50, z-score = -3.92, R) >droSec1.super_7 427082 98 - 3727775 GAACCAACACUGUUCUGGGAGCUUCUGGGGAAACUCAUUCAGACGGCGUAGAUUCAUUCAUUUUAUCUUUUU----UUUUUUUGCUUAUGCU-UAAGCGGUUC (((((.((.((((.(((((......((((....))))))))))))).)).......................----.......(((((....-)))))))))) ( -24.80, z-score = -2.55, R) >droSim1.chr2L_random 622706 102 - 909653 GAACCAACACUGUUCUGGGAGCUUCUGGGGAAACUCAUUCAGACGGCGUAGGUUCAUUCAUUUUUUCUUUUUUUCUUUUUUUUGCAUAUGCU-UAAGCGGUUC (((((.((.((((.(((((......((((....))))))))))))).)).))))).................................(((.-...))).... ( -24.10, z-score = -2.01, R) >consensus GAACCAACACUGUUCUGGGAGCUUCUGGGGAAACUCAUUCAGACGGCGUAGGUUCAUUCAUUUUUUCUU__U____UUUUUUUGCUUAUGCU_UAAGCGGUUC (((((....((((.(((....((..((((....))))...)).))).))))................................(((((.....)))))))))) ( -7.99 = -9.63 + 1.64)

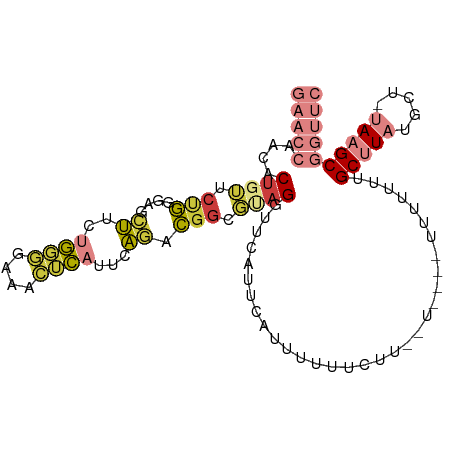
| Location | 16,780,380 – 16,780,478 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 69.94 |
| Shannon entropy | 0.59365 |
| G+C content | 0.46459 |
| Mean single sequence MFE | -28.51 |
| Consensus MFE | -9.59 |
| Energy contribution | -11.56 |
| Covariance contribution | 1.97 |
| Combinations/Pair | 1.52 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.61 |
| SVM RNA-class probability | 0.993330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16780380 98 + 23011544 ---AUGAAAAAAUGAAUGAACC--UACGCCGUCUGAAUGAGUUUCCCCAGAAGCUC-CCAGAACAGUGUUGGUUCCGCUUAGUGCACGCGCUCUUCA-GUAGAUG ---.((((.........(((((--.((((..((((...(((((((....)))))))-.))))...)))).))))).....((((....)))).))))-....... ( -27.30, z-score = -2.79, R) >droGri2.scaffold_15252 13262408 104 + 17193109 AACAAUCUACAAAUUAUGAGCUGAUCGGUAGUGGGAGUAUUUCCUUAAAAAAACUCUCCCGAUCAGU-UUGUAAAUGCUUAAAGCACGCGCUCCUUAUGCAAAAU .............(((..((((((((((....((((((.(((......))).)))))))))))))))-)..))).(((.((((((....)))..))).))).... ( -30.60, z-score = -4.61, R) >droVir3.scaffold_12963 10128149 90 + 20206255 -------AAAAAUUUGUUUAGC--UGGGCCAAAAGGUUGG--CUCCAACCAACUUGUCUGGCCCAGCUCU---UUUGCUUAAAGCACGCGCAAUUUUUGUAUGU- -------.((((.((((..(((--(((((((..(((((((--......)))))))...))))))))))..---..(((.....)))...)))).))))......- ( -33.80, z-score = -3.83, R) >droYak2.chr2R 16817479 97 - 21139217 ----AAAAAAAAUGAAUGAACC--UCCGCCGUCUGAAUGAGUUUCCCCAGAAGCUC-CCAGAACAGUGUGGGUUCCGCUUAGUGCACGCGCUCUUCA-GUAGAUG ----........((((.(((((--..(((..((((...(((((((....)))))))-.))))...)))..))))).....((((....)))).))))-....... ( -27.60, z-score = -2.76, R) >droEre2.scaffold_4845 19851042 98 + 22589142 ---GAAAGAAAAUGAUUGAACC--UCCGCCGUCUGAUUGAGCUUCCCCCGAAGCUC-CCAGAACAGUGUAGGUUCCGCUUAGUGCACGCGCUCUUCA-GUAGAUG ---....(((.......(((((--(.(((..((((...(((((((....)))))))-.))))...))).)))))).....((((....)))).))).-....... ( -28.70, z-score = -2.81, R) >droSec1.super_7 427111 98 + 3727775 ---AAGAUAAAAUGAAUGAAUC--UACGCCGUCUGAAUGAGUUUCCCCAGAAGCUC-CCAGAACAGUGUUGGUUCCGCUUAGUGCACGCGCUCUUCA-GUAGAUG ---.........((((.(((((--.((((..((((...(((((((....)))))))-.))))...)))).))))).....((((....)))).))))-....... ( -24.50, z-score = -1.67, R) >droSim1.chr2L_random 622739 98 + 909653 ---AAGAAAAAAUGAAUGAACC--UACGCCGUCUGAAUGAGUUUCCCCAGAAGCUC-CCAGAACAGUGUUGGUUCCGCUUAGUGCACGCGCUCUUCA-GUAGAUG ---.........((((.(((((--.((((..((((...(((((((....)))))))-.))))...)))).))))).....((((....)))).))))-....... ( -27.10, z-score = -2.70, R) >consensus ___AAGAAAAAAUGAAUGAACC__UACGCCGUCUGAAUGAGUUUCCCCAGAAGCUC_CCAGAACAGUGUUGGUUCCGCUUAGUGCACGCGCUCUUCA_GUAGAUG .................(((((....(((..((((...(((((((....)))))))..))))...)))..))))).((.....)).................... ( -9.59 = -11.56 + 1.97)
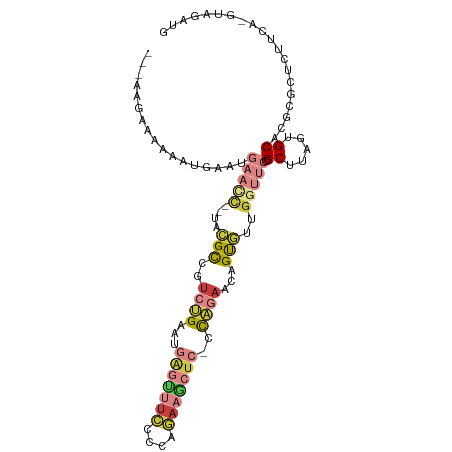
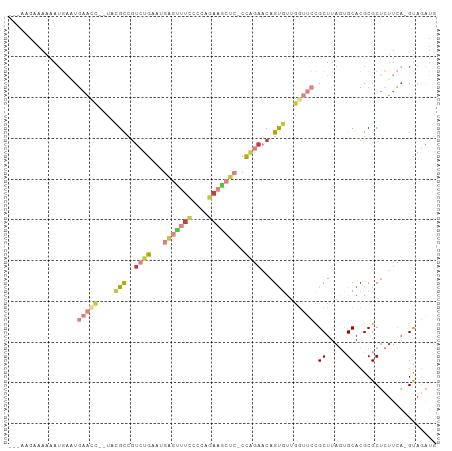
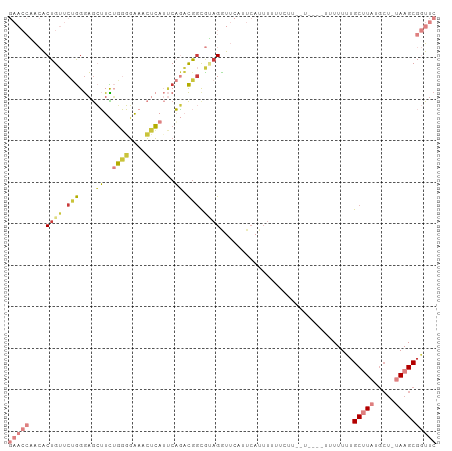
| Location | 16,780,380 – 16,780,478 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 69.94 |
| Shannon entropy | 0.59365 |
| G+C content | 0.46459 |
| Mean single sequence MFE | -28.97 |
| Consensus MFE | -8.94 |
| Energy contribution | -8.93 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.638528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16780380 98 - 23011544 CAUCUAC-UGAAGAGCGCGUGCACUAAGCGGAACCAACACUGUUCUGG-GAGCUUCUGGGGAAACUCAUUCAGACGGCGUA--GGUUCAUUCAUUUUUUCAU--- .......-((((((((((.........)))(((((.((.((((.((((-(......((((....))))))))))))).)).--)))))......))))))).--- ( -29.40, z-score = -2.16, R) >droGri2.scaffold_15252 13262408 104 - 17193109 AUUUUGCAUAAGGAGCGCGUGCUUUAAGCAUUUACAA-ACUGAUCGGGAGAGUUUUUUUAAGGAAAUACUCCCACUACCGAUCAGCUCAUAAUUUGUAGAUUGUU ..((((((....((((..((((.....))))......-..(((((((..((((.((((....)))).))))......)))))))))))......))))))..... ( -27.70, z-score = -2.62, R) >droVir3.scaffold_12963 10128149 90 - 20206255 -ACAUACAAAAAUUGCGCGUGCUUUAAGCAAA---AGAGCUGGGCCAGACAAGUUGGUUGGAG--CCAACCUUUUGGCCCA--GCUAAACAAAUUUUU------- -......(((((((.(...(((.....)))..---.)((((((((((((...((((((....)--)))))..)))))))))--))).....)))))))------- ( -34.30, z-score = -3.90, R) >droYak2.chr2R 16817479 97 + 21139217 CAUCUAC-UGAAGAGCGCGUGCACUAAGCGGAACCCACACUGUUCUGG-GAGCUUCUGGGGAAACUCAUUCAGACGGCGGA--GGUUCAUUCAUUUUUUUU---- .......-((((...(((.........)))(((((..(.((((.((((-(......((((....))))))))))))).)..--))))).))))........---- ( -26.90, z-score = -0.88, R) >droEre2.scaffold_4845 19851042 98 - 22589142 CAUCUAC-UGAAGAGCGCGUGCACUAAGCGGAACCUACACUGUUCUGG-GAGCUUCGGGGGAAGCUCAAUCAGACGGCGGA--GGUUCAAUCAUUUUCUUUC--- .......-.(((((((((.........)))((((((.(.((((.(((.-(((((((....)))))))...))))))).).)--))))).......)))))).--- ( -33.90, z-score = -2.56, R) >droSec1.super_7 427111 98 - 3727775 CAUCUAC-UGAAGAGCGCGUGCACUAAGCGGAACCAACACUGUUCUGG-GAGCUUCUGGGGAAACUCAUUCAGACGGCGUA--GAUUCAUUCAUUUUAUCUU--- .((((((-.((((..(.(..((.....))(((((.......)))))).-)..))))((((....))))..........)))--)))................--- ( -22.30, z-score = -0.02, R) >droSim1.chr2L_random 622739 98 - 909653 CAUCUAC-UGAAGAGCGCGUGCACUAAGCGGAACCAACACUGUUCUGG-GAGCUUCUGGGGAAACUCAUUCAGACGGCGUA--GGUUCAUUCAUUUUUUCUU--- .......-.(((((((((.........)))(((((.((.((((.((((-(......((((....))))))))))))).)).--)))))......))))))..--- ( -28.30, z-score = -1.62, R) >consensus CAUCUAC_UGAAGAGCGCGUGCACUAAGCGGAACCAACACUGUUCUGG_GAGCUUCUGGGGAAACUCAUUCAGACGGCGGA__GGUUCAUUCAUUUUUUCUU___ ............((((...(((.....))).........(((.((((..(((.(((....))).)))...))))))).......))))................. ( -8.94 = -8.93 + -0.01)

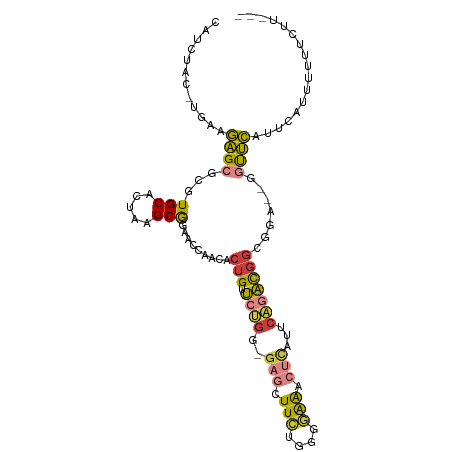
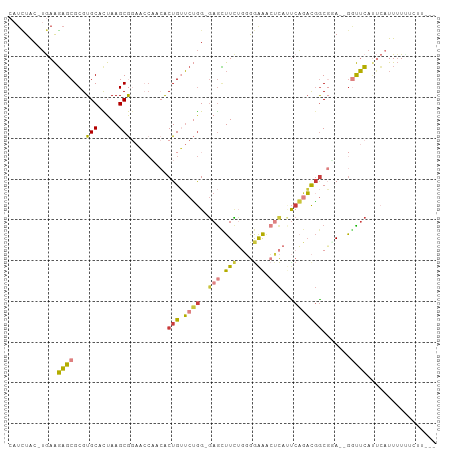
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:27 2011