
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,637,499 – 1,637,603 |
| Length | 104 |
| Max. P | 0.571962 |

| Location | 1,637,499 – 1,637,603 |
|---|---|
| Length | 104 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 76.13 |
| Shannon entropy | 0.43401 |
| G+C content | 0.39881 |
| Mean single sequence MFE | -24.20 |
| Consensus MFE | -11.11 |
| Energy contribution | -10.87 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571962 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1637499 104 - 23011544 --GAUCAUAAAAGUCGACCAAACAAAAGCUUCAGUUAAUCGAAUGGCGCAUUAGUUGAGGUUCGGAUAUUGAUAUCUGAACACCU-GAAAAUAA-----AAAACGACUCACG --.........(((((.............((((..((((((.....)).))))..))))(((((((((....)))))))))....-........-----....))))).... ( -21.80, z-score = -1.83, R) >droPer1.super_8 3331474 111 - 3966273 CAAACCAUCAAAUUAAAUCAAACAAUGGCUUCGGUUAAUCGAAUGGCGCAUUAAGAUAAAUGUGGAUACCGAUAUCGAUGCCGAU-GCCGAUUUUCCCAGGGCAAACUCACG .........................((((.(((((..(((((.(((((((((......))))))....)))...)))))))))).-)))).........(((....)))... ( -23.80, z-score = -0.35, R) >dp4.chr4_group2 675538 111 + 1235136 CAAACCAUCAAAUUAAAUCAAACAAUGGCUUCGGUUAAUCGAAUGGCGCAUUAAGAUAAAUGUGGAUACCGAUAUCGAUGCCGAU-GCCGAUAUUCCCAGGGCAAACUCACG .........................((((.(((((..(((((.(((((((((......))))))....)))...)))))))))).-)))).........(((....)))... ( -23.80, z-score = -0.41, R) >droEre2.scaffold_4929 1686227 105 - 26641161 --GAUCAUAAAAGUCGGCCAAACAAAAGCUUCAGUUAAUCGAAUGGCGCAUUAGGUGAAGUUCAGAUACUGAUAUCUGAACACCUGAAAAAUCA-----AAAACGACUCACG --.........(((((((((......(((....))).......))))...((((((...(((((((((....))))))))))))))).......-----....))))).... ( -27.02, z-score = -3.63, R) >droYak2.chr2L 1612185 110 - 22324452 --GAUCAUAAAAGUCGGCCAAACAAAAGCGUCAGUUAAUCGAAUGGCGCAUUAGCUGAAGUUCAGAUACUGAUAUCUGAACACCUCAAAAAUGAAAUGGAAAACAACUCACG --..........((...(((..........(((((((((((.....)).))))))))).(((((((((....)))))))))...............)))...))........ ( -25.00, z-score = -2.54, R) >droSec1.super_14 1580221 104 - 2068291 --GAUCAUAAAAGUCGGCCAAACAAUAGCUUCAGUUAAUCGAAUGGCGCAUUAGUUGAGGUUCGGAUAUUGAUAUCUGAACACCC-GAAAAUAA-----AAAACGACUCACG --.........(((((((((.....((((....))))......))))...(((.(((.((((((((((....))))))))).).)-))...)))-----....))))).... ( -24.00, z-score = -2.09, R) >droSim1.chr2L 1608175 104 - 22036055 --GAUCAUAAAAGUCGGCCAAACAAUAGCUUCAGUUAAUCGAAUGGCGCAUUAGUUGAGGUUCGGAUAUUGAUAUCUGAACACCC-GAAAAUAA-----AAAACGACUCACG --.........(((((((((.....((((....))))......))))...(((.(((.((((((((((....))))))))).).)-))...)))-----....))))).... ( -24.00, z-score = -2.09, R) >consensus __GAUCAUAAAAGUCGGCCAAACAAUAGCUUCAGUUAAUCGAAUGGCGCAUUAGGUGAAGUUCGGAUACUGAUAUCUGAACACCU_GAAAAUAA_____AAAACGACUCACG .............................((((.(((((((.....)).))))).))))(((((((((....)))))))))............................... (-11.11 = -10.87 + -0.24)

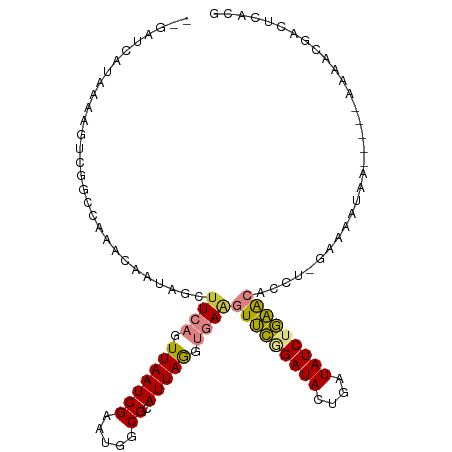

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:49 2011