
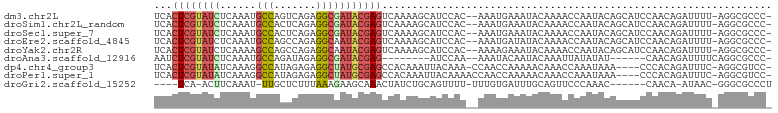
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,778,674 – 16,778,773 |
| Length | 99 |
| Max. P | 0.913386 |

| Location | 16,778,674 – 16,778,773 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 71.09 |
| Shannon entropy | 0.60269 |
| G+C content | 0.41999 |
| Mean single sequence MFE | -13.10 |
| Consensus MFE | -8.96 |
| Energy contribution | -9.12 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.75 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913386 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
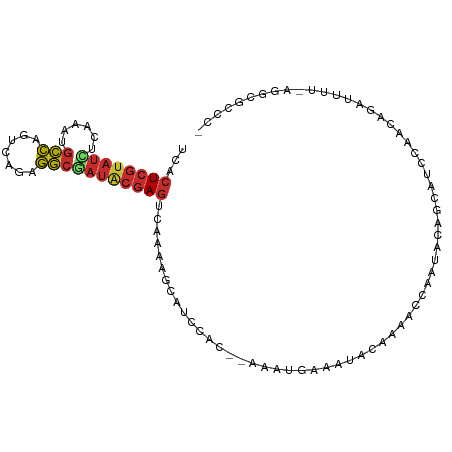
>dm3.chr2L 16778674 99 + 23011544 UCACUCGUAUCUCAAAUGCCAGUCAGAGGCGAUACGAGUCAAAAGCAUCCAC--AAAUGAAAUACAAAACCAAUACAGCAUCCAACAGAUUUU-AGGCGCCC- ..(((((((((......(((.......)))))))))))).............--.......................((..((..........-.)).))..- ( -14.30, z-score = -0.82, R) >droSim1.chr2L_random 620770 99 + 909653 UCACUCGUAUCUCAAAUGCCACUCAGAGGCGAUACGAGUCAAAAGCAUCCAC--AAAUGAAAUACAAAACCAAUACAGCAUCCAACAGAUUUU-AGGCGCCC- ..(((((((((......(((.......)))))))))))).............--.......................((..((..........-.)).))..- ( -14.30, z-score = -1.01, R) >droSec1.super_7 425412 99 + 3727775 UCACUCGUAUCUCAAAUGCCACUCAGAGGCGAUACGAGUCAAAAGCAUCCAC--AAAUGAAAUACAAAACCAAUACAGCAUCCAACAGAUUUU-AGGCGCCC- ..(((((((((......(((.......)))))))))))).............--.......................((..((..........-.)).))..- ( -14.30, z-score = -1.01, R) >droEre2.scaffold_4845 19849339 99 + 22589142 UCACUCGUAUCUCAAAUGCCAGCCAGAGGCAAUACGAGUCAAAAGCAUCCAC--AAAUGAUAUACAAAACCAAUACAGCAUCCAACAGAUUUU-AGGCGCCC- ..((((((((......((((.......)))))))))))).............--.......................((..((..........-.)).))..- ( -13.00, z-score = -0.60, R) >droYak2.chr2R 16815807 99 - 21139217 UCACUCGUAUCUCAAAAGCCAGCCAGAGGCAAUACGAGUCAAAAGCAUCCAC--AAAAGAAAUACAAAACCAAUACAGCAUCCAACAGAUUUU-AGGCGCCC- ..((((((((.......(((.......))).)))))))).............--.......................((..((..........-.)).))..- ( -10.30, z-score = -0.33, R) >droAna3.scaffold_12916 7338380 86 + 16180835 AAUCUCGUAUCUCAAAUGCCAGAUAGAGGCGAUACGAG--------AUCCAA--AAAUACAAUACAAAUUAUAUAU------CAACAGAUUUUCAGGCGCCC- .((((((((((......(((.......)))))))))))--------))....--......................------....................- ( -15.20, z-score = -1.92, R) >dp4.chr4_group3 8690319 96 - 11692001 UCACUCGUAUAUCAAAGGCCAUAGAGAGGCUAUGCGAGCCACAAAUUACAAA-CCAACCAAAAACAAACCAAAUAAA----CCCACAGAUUUC-AGGCGUCC- ....((((((......((((.......))))))))))(((..(((((.....-........................----......))))).-.)))....- ( -11.65, z-score = -0.56, R) >droPer1.super_1 5794660 97 - 10282868 UCACUCGUAUAUCAAAGGCCAUAGAGAGGCUAUGCGAGCCACAAAUUACAAAACCAACCAAAAACAAACCAAAUAAA----CCCACAGAUUUC-AGGCGUCC- ....((((((......((((.......))))))))))(((..(((((..............................----......))))).-.)))....- ( -11.62, z-score = -0.62, R) >droGri2.scaffold_15252 13260600 88 + 17193109 ----UCA-ACUUCAAAU-UUGCUCUUUAAAGAAGCAAACUAUCUGCAGUUUU-UUUGUGAUUUGCAGUUCCCAAAC------CAACA-AUAAC-GGGCGCCCU ----...-........(-(((((((....)).))))))....((((((.((.-.....)).)))))).........------.....-.....-((....)). ( -13.20, z-score = 0.19, R) >consensus UCACUCGUAUCUCAAAUGCCAGUCAGAGGCGAUACGAGUCAAAAGCAUCCAC__AAAUGAAAUACAAAACCAAUACAGCAUCCAACAGAUUUU_AGGCGCCC_ ...((((((((......(((.......)))))))))))................................................................. ( -8.96 = -9.12 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:23 2011