| Sequence ID | dm3.chr2L |
|---|---|
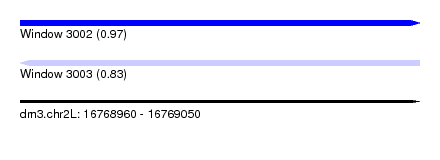
| Location | 16,768,960 – 16,769,050 |
| Length | 90 |
| Max. P | 0.967179 |

| Location | 16,768,960 – 16,769,050 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 51.05 |
| Shannon entropy | 0.95674 |
| G+C content | 0.36888 |
| Mean single sequence MFE | -23.37 |
| Consensus MFE | 0.00 |
| Energy contribution | 0.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 0.00 |
| Mean z-score | -3.47 |
| Structure conservation index | -0.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967179 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
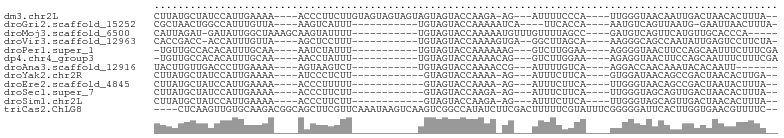
>dm3.chr2L 16768960 90 + 23011544 --UAAAGUGUUAGUCAAUUGUUACCCAA----UGGGAAAAU---CU-UCUUGGUACUACUACUACUACUACAAGAAGGGU----UUUUCAAUGGAUAGCAUAAG --................(((((.(((.----((..(((..---((-(((((..................)))))))..)----))..)).))).))))).... ( -22.67, z-score = -2.23, R) >droGri2.scaffold_15252 13249806 79 + 17193109 -UAAAGUUAAUUC-CAUUAACUGACAUU----UGGUGAA----UGAUUUUUGGUACUACA-----------AAAUGACUU----UAACAAAUGGCCAGUUAGCG -............-..(((((((.((((----((.((((----....(((((......))-----------)))....))----)).))))))..))))))).. ( -16.90, z-score = -2.16, R) >droMoj3.scaffold_6500 2796072 83 + 32352404 -----UGGGUGCAACAUGAACUGACAUC----GGCUAAAACAAACAUUUUUGGUACUACA-----------AAAAUACUUGCUUUAGCCAAUAUC-AUCUAAUG -----..((((((........)).))))----(((((((.(((..(((((((......))-----------)))))..))).)))))))......-........ ( -20.30, z-score = -4.29, R) >droVir3.scaffold_12963 10115757 81 + 20206255 -UAGAAGGACUCAAUAUUGGCUGCCCUU----UGCUAAGCC--UCACUUUUGGUACUACA-----------AAAGGAGCU----UAACAAAUGGU-GGUCGGUG -.............(((((((..((.((----((.(((((.--((.((((((......))-----------)))))))))----)).)))).)).-.))))))) ( -29.30, z-score = -3.61, R) >droPer1.super_1 5784153 81 - 10282868 UCGAAAGAAAUUGCUGGAAGUUACCCCU----UUCCAAGAC---CUUUUUUGGUACUACA-----------AAAUAGAUU----UUGCAAAUGUGUGGCAACA- ((....))..((((((((((......))----)))(((((.---((.(((((......))-----------))).)).))----))).........)))))..- ( -16.00, z-score = -0.76, R) >dp4.chr4_group3 8679984 81 - 11692001 UCGAAAGAAAUUGCUGGAAGUUACCUCU----UUCCAAGAC---CUGUUUUGGUACUACA-----------AAAUAGGUU----UUGCAAAUGUGUGGCAACA- ((....))..((((((((((......))----)))((((((---((((((((......))-----------)))))))))----))).........)))))..- ( -27.00, z-score = -3.89, R) >droAna3.scaffold_12916 7328539 75 + 16180835 -------AAUUGUGUAUUUGUUGGUCCU----UGACAAAAU---CGGUUUUGGUACUACA-----------AGACUUACU----UUUUCAAGGGUCAACAAGUA -------.......(((((((((..(((----(((.(((.(---.(((((((......))-----------))))).).)----)).))))))..))))))))) ( -29.00, z-score = -5.87, R) >droYak2.chr2R 16806171 78 - 21139217 --UCAAGUGUUAGUCGGCUGUUAUCCAC----UGAAGAAAU---CU-UUUUGGUACUAC------------AAGAGGGAU----UUUUCAAUGGAUAGCAUAAG --................(((((((((.----(((((((.(---((-(((((......)------------))))))).)----)))))).))))))))).... ( -26.50, z-score = -4.58, R) >droEre2.scaffold_4845 19839703 78 + 22589142 --UAAAGUAUUAGUCGGCUGUUACCCAA----UGAAGAAAU---CU-UUUUGGUACUAC------------AAAAAGGGU----UUUUCAAUGGAUAGCAUAAG --..............(((((...(((.----(((((((..---((-(((((......)------------))))))..)----)))))).))))))))..... ( -22.70, z-score = -4.03, R) >droSec1.super_7 415768 78 + 3727775 --UAAAGUGUUAGUCAACUGCUACCCAA----UGAAGAAAU---CU-UCUUGGUACUAC------------AAGAAGGGU----UUUUCAAUGGAUAGCAUAAG --...(((........)))((((.(((.----(((((((..---((-(((((......)------------))))))..)----)))))).))).))))..... ( -27.50, z-score = -5.14, R) >droSim1.chr2L 16469937 78 + 22036055 --UAAAGUGUUAGUCAACUGCUACCCAA----UGAAGAAAU---CU-UCUUGGUACUAC------------AAGAAGGGU----UUUUCAAUGGAUAGCAUAAG --...(((........)))((((.(((.----(((((((..---((-(((((......)------------))))))..)----)))))).))).))))..... ( -27.50, z-score = -5.14, R) >triCas2.ChLG8 11594924 98 - 15773733 --GAAAACGUUCACCAAGUGAAUCCCCCGAAAUACGAAAAAGUCGAAGAUAUGGCCGACUUGACUUAUUUGAACGAAGCUGCCGUCUUGCACAACUUGAG---- --.....((((((..((((........((.....))...((((((..........)))))).))))...))))))..(.(((......))))........---- ( -15.10, z-score = 0.08, R) >consensus __UAAAGUAUUAGUCAACUGUUACCCAU____UGAAAAAAU___CU_UUUUGGUACUACA___________AAAAAGGGU____UUUUCAAUGGAUAGCAAAAG ........................................................................................................ ( 0.00 = 0.00 + 0.00)

| Location | 16,768,960 – 16,769,050 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 51.05 |
| Shannon entropy | 0.95674 |
| G+C content | 0.36888 |
| Mean single sequence MFE | -20.68 |
| Consensus MFE | 0.00 |
| Energy contribution | 0.00 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 0.00 |
| Mean z-score | -2.96 |
| Structure conservation index | -0.00 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.82 |
| SVM RNA-class probability | 0.826160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16768960 90 - 23011544 CUUAUGCUAUCCAUUGAAAA----ACCCUUCUUGUAGUAGUAGUAGUAGUACCAAGA-AG---AUUUUCCCA----UUGGGUAACAAUUGACUAACACUUUA-- ....((.((((((.((..((----(..(((((((..(((.((....)).))))))))-))---..)))..))----.)))))).))................-- ( -18.50, z-score = -1.46, R) >droGri2.scaffold_15252 13249806 79 - 17193109 CGCUAACUGGCCAUUUGUUA----AAGUCAUUU-----------UGUAGUACCAAAAAUCA----UUCACCA----AAUGUCAGUUAAUG-GAAUUAACUUUA- ((.((((((((.(((((...----......(((-----------((......)))))....----.....))----))))))))))).))-............- ( -13.47, z-score = -1.25, R) >droMoj3.scaffold_6500 2796072 83 - 32352404 CAUUAGAU-GAUAUUGGCUAAAGCAAGUAUUUU-----------UGUAGUACCAAAAAUGUUUGUUUUAGCC----GAUGUCAGUUCAUGUUGCACCCA----- (((....(-((((((((((((((((((((((((-----------((......))))))))))))))))))))----)))))))....))).........----- ( -32.60, z-score = -7.46, R) >droVir3.scaffold_12963 10115757 81 - 20206255 CACCGACC-ACCAUUUGUUA----AGCUCCUUU-----------UGUAGUACCAAAAGUGA--GGCUUAGCA----AAGGGCAGCCAAUAUUGAGUCCUUCUA- ....((((-(...(((((((----(((((((((-----------((......)))))).))--.))))))))----))((....)).....)).)))......- ( -21.90, z-score = -2.12, R) >droPer1.super_1 5784153 81 + 10282868 -UGUUGCCACACAUUUGCAA----AAUCUAUUU-----------UGUAGUACCAAAAAAG---GUCUUGGAA----AGGGGUAACUUCCAGCAAUUUCUUUCGA -.((((((.(...(((.(((----.((((.(((-----------((......))))).))---)).))).))----).)))))))................... ( -17.20, z-score = -0.95, R) >dp4.chr4_group3 8679984 81 + 11692001 -UGUUGCCACACAUUUGCAA----AACCUAUUU-----------UGUAGUACCAAAACAG---GUCUUGGAA----AGAGGUAACUUCCAGCAAUUUCUUUCGA -.((((((.....(((.(((----.((((.(((-----------((......))))).))---)).))).))----)..))))))................... ( -18.20, z-score = -1.50, R) >droAna3.scaffold_12916 7328539 75 - 16180835 UACUUGUUGACCCUUGAAAA----AGUAAGUCU-----------UGUAGUACCAAAACCG---AUUUUGUCA----AGGACCAACAAAUACACAAUU------- ...((((((..((((((.((----(((..((.(-----------((......))).))..---))))).)))----)))..))))))..........------- ( -18.80, z-score = -3.53, R) >droYak2.chr2R 16806171 78 + 21139217 CUUAUGCUAUCCAUUGAAAA----AUCCCUCUU------------GUAGUACCAAAA-AG---AUUUCUUCA----GUGGAUAACAGCCGACUAACACUUGA-- ....((.(((((((((((((----(((....((------------(......)))..-.)---)))).))))----))))))).))................-- ( -16.80, z-score = -2.98, R) >droEre2.scaffold_4845 19839703 78 - 22589142 CUUAUGCUAUCCAUUGAAAA----ACCCUUUUU------------GUAGUACCAAAA-AG---AUUUCUUCA----UUGGGUAACAGCCGACUAAUACUUUA-- ....((.((((((.((((.(----(..((((((------------(......)))))-))---..)).))))----.)))))).))................-- ( -16.50, z-score = -2.96, R) >droSec1.super_7 415768 78 - 3727775 CUUAUGCUAUCCAUUGAAAA----ACCCUUCUU------------GUAGUACCAAGA-AG---AUUUCUUCA----UUGGGUAGCAGUUGACUAACACUUUA-- ....(((((((((.((((.(----(..((((((------------(......)))))-))---..)).))))----.)))))))))................-- ( -25.80, z-score = -5.25, R) >droSim1.chr2L 16469937 78 - 22036055 CUUAUGCUAUCCAUUGAAAA----ACCCUUCUU------------GUAGUACCAAGA-AG---AUUUCUUCA----UUGGGUAGCAGUUGACUAACACUUUA-- ....(((((((((.((((.(----(..((((((------------(......)))))-))---..)).))))----.)))))))))................-- ( -25.80, z-score = -5.25, R) >triCas2.ChLG8 11594924 98 + 15773733 ----CUCAAGUUGUGCAAGACGGCAGCUUCGUUCAAAUAAGUCAAGUCGGCCAUAUCUUCGACUUUUUCGUAUUUCGGGGGAUUCACUUGGUGAACGUUUUC-- ----...(((((((........)))))))((((((..(((((...(((((........)))))(((..((.....))..)))...))))).)))))).....-- ( -22.60, z-score = -0.76, R) >consensus CUUAUGCUAUCCAUUGAAAA____AACCUUUUU___________UGUAGUACCAAAA_AG___AUUUUUGCA____AUGGGUAACAAAUGACUAACACUUUA__ ........................................................................................................ ( 0.00 = 0.00 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:19 2011