| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,752,687 – 16,752,779 |
| Length | 92 |
| Max. P | 0.926785 |

| Location | 16,752,687 – 16,752,779 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 66.91 |
| Shannon entropy | 0.67907 |
| G+C content | 0.43970 |
| Mean single sequence MFE | -22.36 |
| Consensus MFE | -8.16 |
| Energy contribution | -8.26 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.33 |
| SVM RNA-class probability | 0.926785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
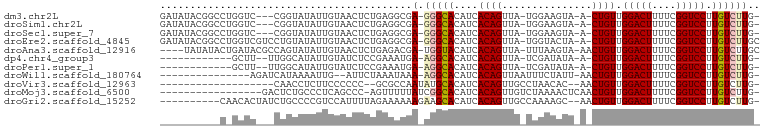
>dm3.chr2L 16752687 92 + 23011544 GAUAUACGGCCUGGUC---CGGUAUAUUGUAACUCUGAGGCGA-GGGCACAUCACAGUUA-UGGAAGUA-A-CUGUUGGACUUUUCGGUCCUUGUCUUG- (((((((((......)---).)))))))........(((((((-((((.((..(((((((-(....)))-)-))))))((....)).))))))))))).- ( -28.70, z-score = -1.62, R) >droSim1.chr2L 16451847 92 + 22036055 GAUAUACGGCCUGGUC---CGGUAUAUUGUAACUCUGAGGCGA-GGGCACAUCACAGUUA-UGGAAGUA-A-CUGUUGGACUUUUCGGUCCUUGUCUUG- (((((((((......)---).)))))))........(((((((-((((.((..(((((((-(....)))-)-))))))((....)).))))))))))).- ( -28.70, z-score = -1.62, R) >droSec1.super_7 399503 92 + 3727775 GAUAUACGGCCUGGUC---CGGUAUAUUGUAACUCUGAGGCGA-GGGCACAUCACAGUUA-UGGAAGUA-A-CUGUUGGACUUUUCGGUCCUUGUCUUG- (((((((((......)---).)))))))........(((((((-((((.((..(((((((-(....)))-)-))))))((....)).))))))))))).- ( -28.70, z-score = -1.62, R) >droEre2.scaffold_4845 19824431 96 + 22589142 GAUAUACGGCCUGGUCGUCCUGUAUAUUGUAACUCUGAGGCGA-GGGCACAUCACAGUUA-UGGUACUA-A-CUGUUGGACUUUUCGGUCCUUGUCUUGC (((((((((..........)))))))))........(((((((-((((.((..(((((((-......))-)-))))))((....)).))))))))))).. ( -27.70, z-score = -1.01, R) >droAna3.scaffold_12916 7310203 93 + 16180835 ----UAUAUACUGAUACGCCAGUAUAUUGUAACUCUGAGACGA-UGGUACAUCACAGUUA-UUUAAGUA-AACUGUUGGACUUUUCGGUCCUUGUCUUGC ----.((((((((......)))))))).........(((((((-((...))))((((((.-........-)))))).(((((....)))))..))))).. ( -23.30, z-score = -1.78, R) >dp4.chr4_group3 8661564 81 - 11692001 ------------GCUU--UUGGCAUAUUGUAUCUCCGAAAUGA-AGGCACAUCACAGUUA-UCGAUAUA-A-CUGUUGGACUUUUCGGUCCUUGUCUUG- ------------..((--((((............))))))..(-(((((....(((((((-(....)))-)-)))).(((((....))))).)))))).- ( -19.00, z-score = -1.71, R) >droPer1.super_1 5765438 81 - 10282868 ------------GCUU--UUGGCAUAUUGUAUCUCCGAAAUGA-AGGCACAUCACAGUUA-UCGAUAUA-A-CUGUUGGACUUUUCGGUCCUUGUCUUG- ------------..((--((((............))))))..(-(((((....(((((((-(....)))-)-)))).(((((....))))).)))))).- ( -19.00, z-score = -1.71, R) >droWil1.scaffold_180764 1100678 80 + 3949147 ---------------AGAUCAUAAAAUUG--AUUCUAAAUAAA-AGGCACAUCACAGUUAAUUUCUAUU-AACUGUUGGACUUUUCGGUCCUUGUCUUG- ---------------.(((((......))--)))........(-(((((....(((((((((....)))-)))))).(((((....))))).)))))).- ( -21.30, z-score = -4.44, R) >droVir3.scaffold_12963 10095950 76 + 20206255 -------------------CAACCUCUUCCCCCC--GCGCCAAUAUGCACAUCACAGUUGCCUAACAC--AACUGUUGGACUUUUCGGUCCUUGUCUUG- -------------------...............--(((......))).....(((((((.......)--)))))).(((((....)))))........- ( -13.90, z-score = -1.60, R) >droMoj3.scaffold_6500 2774372 81 + 32352404 -----------------GACUCUGCCCUCAGCCC-AGUUUUUAUCGGCACAUCACAGUUGUCUAAAACUCAACUGUUGGACUUUUCGGUCCUUGUCUUG- -----------------((((..((.....))..-))))......((((....(((((((.........))))))).(((((....))))).))))...- ( -17.40, z-score = -1.54, R) >droGri2.scaffold_15252 13230375 87 + 17193109 ----------CAACACUAUCUGCCCCGUCCAUUUUAGAAAAAAGAAGCACAUCACAGUUGCCAAAAGC--AACUGUUGGACUUUUCGGUCCUUGUCUUG- ----------..(((....(((....(((((............((......))((((((((.....))--)))))))))))....)))....)))....- ( -18.30, z-score = -1.34, R) >consensus __________C_GGUC___CGGUAUAUUGUAACUCUGAGACGA_AGGCACAUCACAGUUA_UGGAAAUA_A_CUGUUGGACUUUUCGGUCCUUGUCUUG_ .............................................((((....((((...............)))).(((((....))))).)))).... ( -8.16 = -8.26 + 0.10)
| Location | 16,752,687 – 16,752,779 |
|---|---|
| Length | 92 |
| Sequences | 11 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 66.91 |
| Shannon entropy | 0.67907 |
| G+C content | 0.43970 |
| Mean single sequence MFE | -20.88 |
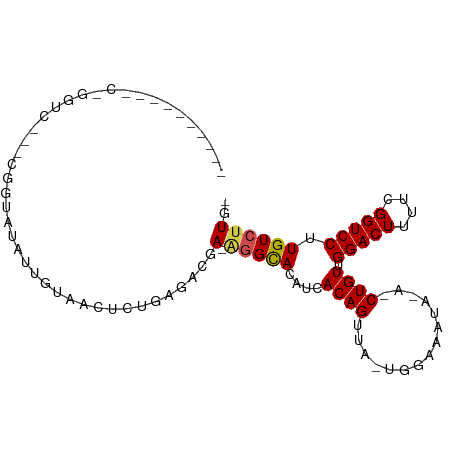
| Consensus MFE | -7.12 |
| Energy contribution | -7.12 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.915444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16752687 92 - 23011544 -CAAGACAAGGACCGAAAAGUCCAACAG-U-UACUUCCA-UAACUGUGAUGUGCCC-UCGCCUCAGAGUUACAAUAUACCG---GACCAGGCCGUAUAUC -........((.((.....((((.((((-(-((......-)))))))..((((..(-((......))).)))).......)---)))..))))....... ( -21.00, z-score = -1.55, R) >droSim1.chr2L 16451847 92 - 22036055 -CAAGACAAGGACCGAAAAGUCCAACAG-U-UACUUCCA-UAACUGUGAUGUGCCC-UCGCCUCAGAGUUACAAUAUACCG---GACCAGGCCGUAUAUC -........((.((.....((((.((((-(-((......-)))))))..((((..(-((......))).)))).......)---)))..))))....... ( -21.00, z-score = -1.55, R) >droSec1.super_7 399503 92 - 3727775 -CAAGACAAGGACCGAAAAGUCCAACAG-U-UACUUCCA-UAACUGUGAUGUGCCC-UCGCCUCAGAGUUACAAUAUACCG---GACCAGGCCGUAUAUC -........((.((.....((((.((((-(-((......-)))))))..((((..(-((......))).)))).......)---)))..))))....... ( -21.00, z-score = -1.55, R) >droEre2.scaffold_4845 19824431 96 - 22589142 GCAAGACAAGGACCGAAAAGUCCAACAG-U-UAGUACCA-UAACUGUGAUGUGCCC-UCGCCUCAGAGUUACAAUAUACAGGACGACCAGGCCGUAUAUC .........((.((.....((((.((((-(-((......-)))))))..((((..(-((......))).)))).......)))).....))))....... ( -24.20, z-score = -2.14, R) >droAna3.scaffold_12916 7310203 93 - 16180835 GCAAGACAAGGACCGAAAAGUCCAACAGUU-UACUUAAA-UAACUGUGAUGUACCA-UCGUCUCAGAGUUACAAUAUACUGGCGUAUCAGUAUAUA---- ...((((..((((......)))).((((((-........-.))))))((((...))-))))))..........(((((((((....))))))))).---- ( -22.70, z-score = -2.56, R) >dp4.chr4_group3 8661564 81 + 11692001 -CAAGACAAGGACCGAAAAGUCCAACAG-U-UAUAUCGA-UAACUGUGAUGUGCCU-UCAUUUCGGAGAUACAAUAUGCCAA--AAGC------------ -........((((......)))).((((-(-(((....)-)))))))..((((.((-((.....)))).)))).........--....------------ ( -18.40, z-score = -2.36, R) >droPer1.super_1 5765438 81 + 10282868 -CAAGACAAGGACCGAAAAGUCCAACAG-U-UAUAUCGA-UAACUGUGAUGUGCCU-UCAUUUCGGAGAUACAAUAUGCCAA--AAGC------------ -........((((......)))).((((-(-(((....)-)))))))..((((.((-((.....)))).)))).........--....------------ ( -18.40, z-score = -2.36, R) >droWil1.scaffold_180764 1100678 80 - 3949147 -CAAGACAAGGACCGAAAAGUCCAACAGUU-AAUAGAAAUUAACUGUGAUGUGCCU-UUUAUUUAGAAU--CAAUUUUAUGAUCU--------------- -....(((.((((......)))).((((((-(((....)))))))))..)))....-.......(((.(--((......))))))--------------- ( -15.40, z-score = -2.01, R) >droVir3.scaffold_12963 10095950 76 - 20206255 -CAAGACAAGGACCGAAAAGUCCAACAGUU--GUGUUAGGCAACUGUGAUGUGCAUAUUGGCGC--GGGGGGAAGAGGUUG------------------- -.........((((......(((.((((((--((.....))))))))..(((((......))))--)...)))...)))).------------------- ( -22.40, z-score = -2.05, R) >droMoj3.scaffold_6500 2774372 81 - 32352404 -CAAGACAAGGACCGAAAAGUCCAACAGUUGAGUUUUAGACAACUGUGAUGUGCCGAUAAAAACU-GGGCUGAGGGCAGAGUC----------------- -...(((..((((......))))..(((((.((((((.....((......)).......))))))-.)))))........)))----------------- ( -18.50, z-score = -0.66, R) >droGri2.scaffold_15252 13230375 87 - 17193109 -CAAGACAAGGACCGAAAAGUCCAACAGUU--GCUUUUGGCAACUGUGAUGUGCUUCUUUUUUCUAAAAUGGACGGGGCAGAUAGUGUUG---------- -(((.((..((((......)))).((((((--((.....)))))))).((.((((((....(((......))).)))))).)).)).)))---------- ( -26.70, z-score = -2.16, R) >consensus _CAAGACAAGGACCGAAAAGUCCAACAG_U_UACUUCAA_UAACUGUGAUGUGCCU_UCGCCUCAGAGUUACAAUAUACCG___GACC_G__________ .....(((.((((......)))).((((...............))))..)))................................................ ( -7.12 = -7.12 + 0.00)

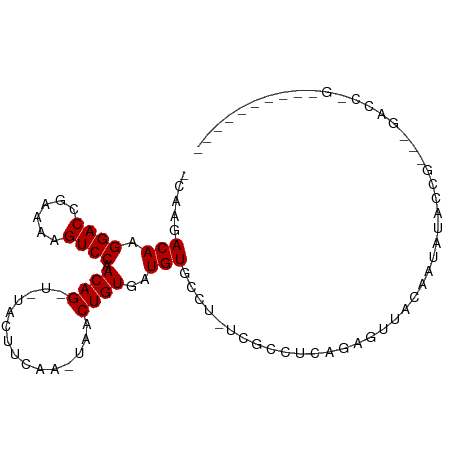
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:17 2011