
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,751,422 – 16,751,531 |
| Length | 109 |
| Max. P | 0.998320 |

| Location | 16,751,422 – 16,751,531 |
|---|---|
| Length | 109 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 69.10 |
| Shannon entropy | 0.68903 |
| G+C content | 0.47954 |
| Mean single sequence MFE | -35.02 |
| Consensus MFE | -14.69 |
| Energy contribution | -14.71 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.32 |
| SVM RNA-class probability | 0.998320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16751422 109 + 23011544 AAAACUGGCCAAAAAGAAGCGGUUGAAUCC---UCCGAGUAUUCACU----UCGCUCGGGCCAGUUUAGCGCAAAAUUGAUGUUGGACUUUUGGGUCUGGCAAAUUAGCCGCACCG .(((((((((.....((.((((.(((((.(---.....).)))))..----)))))).))))))))).(((...((((..(((..((((....))))..)))))))...))).... ( -37.50, z-score = -2.41, R) >droSim1.chr2L 16450602 109 + 22036055 AAAACUGGCCAAAAAGAAGCGGUUGAAUCC---UCCGAGUAUUCACU----UCGCUCUGGCCAGUUUAGCGCAAAAUUGAUGUUGGACUUUUGGGUCUGGCAAAUUAGCCGCACCG .((((((((((....((.((((.(((((.(---.....).)))))..----)))))))))))))))).(((...((((..(((..((((....))))..)))))))...))).... ( -37.30, z-score = -2.67, R) >droSec1.super_7 398256 109 + 3727775 AAAACUGGCCAAAAAGAAGCGGUUGAAUCC---UCCGAGCAUUCACU----UCGCUCGGGCCAGUUUAGCGCAAAAUUGAUGUUGGACUUUUGGGUCUGGCAAAUUAGCCGCACCG .(((((((((..........((......))---..(((((.......----..)))))))))))))).(((...((((..(((..((((....))))..)))))))...))).... ( -37.60, z-score = -2.15, R) >droYak2.chr2R 16787893 106 - 21139217 AAAACUGCCCAAAAAGAUGCGGUUGAAUCUGAGU----AUUCCCC------UCACUCGGGCCAGUUUAGCGCAAAAUUGAUGUUGGACUUUUGGGUCUGGCAAAUUUGCCGCACCG .(((((((((....((((........))))((((----.......------..))))))).)))))).(((((((.....(((..((((....))))..)))..)))).))).... ( -32.40, z-score = -1.50, R) >droEre2.scaffold_4845 19823123 106 + 22589142 AAAACUGGCCAAAAAGCAGCGGUUGAAUCG------GAGUAUUCCCC----UCACUCCGGCCAGUUUAGCGCAAAAUUGAUGUUGGACUUUCGGGUCUGGCAAAUUAGCCGCACCG .(((((((((....(((....))).....(------((((.......----..)))))))))))))).(((...((((..(((..((((....))))..)))))))...))).... ( -35.30, z-score = -1.85, R) >droAna3.scaffold_12916 7309043 97 + 16180835 AAAACUACCCAAACGGUAUCUAACAAAUCC-----------UACA--------AGUUGGGCUAGUUUAGCGCAAAAUUGAUGUUGGACUUUUGGGUCUGGCAAAUUCGCCGCACCG .(((((((((((...(((............-----------))).--------..))))).)))))).(((...((((..(((..((((....))))..)))))))...))).... ( -26.40, z-score = -1.73, R) >dp4.chr4_group3 8659835 115 - 11692001 AAAACUGCCCUAGAAUAU-CGAGUGAAUCCGAAUCUCCAUCCCCCCUCCAUUCACUCGGGGCAGUUUAGCGCAAAAUUGAUGUUGGACUUUGGGGUCUGGCAAAAUUGCCGCACCG .((((((((((.(.....-)((((((((.....................)))))))))))))))))).(((...((((..(((..((((....))))..))).))))..))).... ( -39.10, z-score = -2.93, R) >droPer1.super_1 5763707 115 - 10282868 AAAACUGCCCUAGAAUAU-CGAGUGAAUCCGAAUCUCCAUCCCCCCUUCAUUCACUCGGGGUAGUUUAGCGCAAAAUUGAUGUUGGACUUUGGGGUCUGGCAAAAUUGCCGCACCG .((((((((((.(.....-)((((((((..(((.............))))))))))))))))))))).(((...((((..(((..((((....))))..))).))))..))).... ( -37.82, z-score = -2.78, R) >droWil1.scaffold_180764 1098914 92 + 3949147 ----------AAAAACUACCAGUGGAAUUU----------CUCCAUCAA----UUCUGGUUCAGUUUAGCGCAAAAUUAAUGUUGGACUUUUGAGUCUGGCAAAUUUGCCGCACCG ----------..((((((((((((((....----------.))))....----..)))))..))))).(((((((.....(((..((((....))))..)))..)))).))).... ( -26.10, z-score = -2.95, R) >droVir3.scaffold_12963 10094659 95 + 20206255 AAAACUGCCACUUU-----UGACGGUU-----------UUGUACCG-UUAGUU---UGGCCA-GUUUAGCGCAAAAUUGAUGUUGGACUCUCGGGUCUGGCAUUUUCGCCGCAUCG .(((((((((...(-----(((((((.-----------....))))-))))..---))).))-)))).(((.......(((((..((((....))))..))))).....))).... ( -36.20, z-score = -4.16, R) >droMoj3.scaffold_6500 2773066 103 + 32352404 AAAUCCUCCAGAGUUC---UGAUGGUAAU---------UUGUGCCGCGUAGUC-UCUGGUUGGGUUUAGCGCAAAAUUGAUGUUGGACUCUCGGGUCUGGCAUAUUCGCCGCAUUG ((((((.((((((..(---((.(((((..---------...)))))..))).)-)))))..)))))).(((........((((..((((....))))..))))......))).... ( -36.94, z-score = -3.22, R) >droGri2.scaffold_15252 13229047 108 + 17193109 AAAACUGGCUGGGUAAAAAUGAUGGUAACA-------CUUGUACCG-GUAGCCACCCGGCCACGUUUAGCGCAAAAUUGAUGUUGGACUCUCGGGUCUGGCAAAUUCGCCGCAUCG .(((((((((((((........(((((.(.-------...))))))-......))))))))).)))).(((...((((..(((..((((....))))..)))))))...))).... ( -37.54, z-score = -2.17, R) >consensus AAAACUGCCCAAAAAGAAGCGAUUGAAUCC________GUAUCCCCU____UCACUCGGGCCAGUUUAGCGCAAAAUUGAUGUUGGACUUUUGGGUCUGGCAAAUUAGCCGCACCG ..............................................................(((((((((((....)).)))))))))....(((.((((......)))).))). (-14.69 = -14.71 + 0.01)

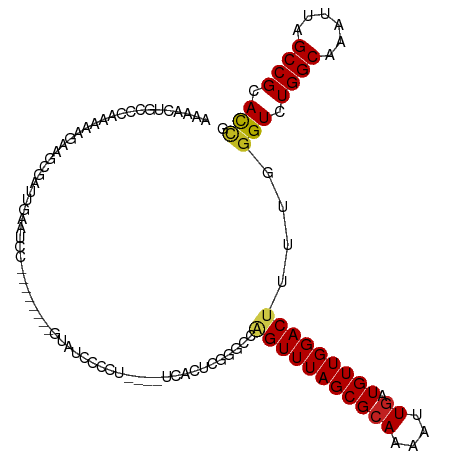
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:16 2011