
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,698,753 – 16,698,882 |
| Length | 129 |
| Max. P | 0.999633 |

| Location | 16,698,753 – 16,698,882 |
|---|---|
| Length | 129 |
| Sequences | 13 |
| Columns | 148 |
| Reading direction | forward |
| Mean pairwise identity | 60.06 |
| Shannon entropy | 0.83426 |
| G+C content | 0.31766 |
| Mean single sequence MFE | -28.41 |
| Consensus MFE | -10.24 |
| Energy contribution | -10.70 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.70 |
| Mean z-score | -2.85 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.11 |
| SVM RNA-class probability | 0.999633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
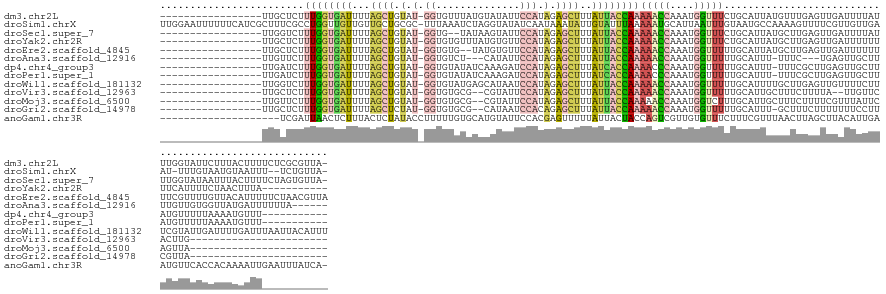
>dm3.chr2L 16698753 129 + 23011544 -----------------UUGCUCUUUGGUGAUUUUAGCUGUAU-GGUGUUUAUGUAUAUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUCUGCAUUAUGUUUGAGUUGAUUUUAUUUGGUAUUCUUUACUUUUCUCGCGUUA- -----------------..((..(((((((((...((((.(((-((.((........)).))))).))))..))))))))).(((((((((..((.((.(((....))))).))..)))))))))................))....- ( -36.00, z-score = -5.08, R) >droSim1.chrX 10558594 143 + 17042790 UUGGAAUUUUUUCAUCGCUUUCGCCUGGUUGUUGUUGCUGCGC-UUUAAAUCUAGGUAUAUCAAUAAAUAUUGUAUUUAAAAAUGCAUUAAUUUGUAAUGCCAAAAGUUUUCGUUGUUGAAU-UUUGUAAUGUAAUUU--UCUGUUA- ..((((..((..(((..(....((((((...(((..((...))-..)))..))))))...(((((((.((.((((((....)))))).)).((((......))))........)))))))..-...)..))).))..)--)))....- ( -23.50, z-score = -0.29, R) >droSec1.super_7 345765 127 + 3727775 -----------------UUGGUCUUUGGUGAUUUUAGCUGUAU-GGUG--UAUAAGUAUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUCUGCAUUAUGCUUGAGUUGAUUUUAUUUGGUAUAAUUUACUUUUCUAGUGUUA- -----------------......(((((((((...((((.(((-((.(--((....))).))))).))))..))))))))).(((((((((..((.((.(((....))))).))..)))))))))......((((.....))))...- ( -35.90, z-score = -4.23, R) >droYak2.chr2R 16734897 119 - 21139217 -----------------UUGCUCUUUGGUGAUUUUAGCUGUAU-GGUGUGUUUAUGUGUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUCUGCAUUAUGCUUGAGUUGAUUUUUUUUCAUUUUCUAACUUUA----------- -----------------..((((.((((((((...((((.(((-((.(..(....)..).))))).))))..))))))))(((((....)))))..((.....))..))))..........................----------- ( -29.30, z-score = -3.34, R) >droEre2.scaffold_4845 19768399 128 + 22589142 -----------------UUGCUCUUUGGUGAUUUUAGCUGUAU-GGUGUG--UAUGUGUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUUUGCAUUAUGCUUGAGUUGAUUUUUUUUCGUUUUGUUACAUUUUUCUAACGUUA -----------------..((((...((((((...((((.(((-((....--........))))).))))..))))))(((((((....)))))))((.....))..)))).................((((........)))).... ( -29.20, z-score = -2.61, R) >droAna3.scaffold_12916 316217 117 - 16180835 -----------------UUGUUCUUUGGUGAUUUUAGCUGUAU-GGUGUCU---CAUAUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUUUGCAUUU-UUUC---UGAGUUGCUUUUGUUGUGGUUAUGAUUUUUUA------ -----------------......(((((((((...((((.(((-((.....---......))))).))))..)))))))))(((((((..(.....(((.((-(...---.))).)))..)..)).)))))...........------ ( -29.00, z-score = -3.45, R) >dp4.chr4_group3 8444203 118 + 11692001 -----------------UUGAUCUUUGGUGAUUUUAGCUGUAU-GGUGUAUAUCAAAGAUCCAUAGAGCUUUAUCACCAAAACCCAAAUGGUUUUUGCAUUU-UUUCGCUUGAGUUGCUUAUGUUUUUAAAAUGUUU----------- -----------------.......((((((((...((((.(((-((..............))))).))))..))))))))((((.....))))...((((((-(...((.((((...)))).))....)))))))..----------- ( -29.14, z-score = -2.75, R) >droPer1.super_1 9895322 118 + 10282868 -----------------UUGAUCUUUGGUGAUUUUAGCUGUAU-GGUGUAUAUCAAAGAUCCAUAGAGCUUUAUCACCAAAACCCAAAUGGUUUUUGCAUUU-UUUCGCUUGAGUUGCUUAUGUUUUUAAAAUGUUU----------- -----------------.......((((((((...((((.(((-((..............))))).))))..))))))))((((.....))))...((((((-(...((.((((...)))).))....)))))))..----------- ( -29.14, z-score = -2.75, R) >droWil1.scaffold_181132 161966 130 - 1035393 -----------------UUGGUCUUUGGUGAUUUUAGCUGUAU-GGUGUAUGAGCAUAAUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUUUGCAUUUUGCUUGAGUUGUUUUCUUUCGUAUUGAUUUUGAUUUAAUUACAUUU -----------------.((((.(((((((((...((((.(((-((..(((....)))..))))).))))..))))))))).))))(((..(((..((.....))..)))..))).......(((((((.......)))).))).... ( -34.80, z-score = -3.78, R) >droVir3.scaffold_12963 7343316 103 - 20206255 -----------------UUGCUCUUUGGUGAUUUUAGCUGUAU-GGUGUGCG--CGUAUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUUUGCAUUGCUUUCUUUUA--UUGUUCACUUG----------------------- -----------------.(((.....((((((...((((.(((-((.(((..--..))).))))).))))..))))))(((((((....)))))))))).............--...........----------------------- ( -29.50, z-score = -3.88, R) >droMoj3.scaffold_6500 7202316 105 + 32352404 -----------------UUGUUCUUUGGUGAUUUUAGCUGUAU-GGUGUGCG--CGUAUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUCUUUGCAUUGCUUUCUUUUCGUUUAUUCAGUUA----------------------- -----------------..(((.(((((((((...((((.(((-((.(((..--..))).))))).))))..)))))))))))).((((((.....((...)).......)))))).........----------------------- ( -25.80, z-score = -2.72, R) >droGri2.scaffold_14978 831307 104 + 1124632 -----------------UUGCUCUUUGGUGAUUUUAGCUCUAU-GGUGUGCG--CAUAAUCCACAGAGCUUUAUUACCAAAAACCAAAUGGUUUUUGCAUUU-GCUUUCUUUUUUUCCUUCGUUA----------------------- -----------------.(((.....((((((...((((((.(-((......--......))).))))))..))))))(((((((....))))))))))...-......................----------------------- ( -27.10, z-score = -3.56, R) >anoGam1.chr3R 47383922 127 - 53272125 --------------------UCGAUUAACUCUUUACUCUAUACCUUUUUGUGCAUGUAUUCCACGAGUUUUUAUUACUACCAGUCGUUGUGUUUCUUUCGUUUAACUUAGCUUACAUUGAAUGUUCACCACAAAAUUGAAUUUAUCA- --------------------(((((((((((..(((..((((......))))...)))......))))).................(((((.......(((((((...........))))))).....)))))))))))........- ( -11.00, z-score = 1.40, R) >consensus _________________UUGCUCUUUGGUGAUUUUAGCUGUAU_GGUGUGUGUACGUAUUCCAUAGAGCUUUAUUACCAAAAACCAAAUGGUUUUUGCAUUUUGUUUGAGUUGAUUUUUUAUGUUUUUAUUAUGUUU___________ ........................((((((((...((((.(...((..............))..).))))..))))))))(((((....)))))...................................................... (-10.24 = -10.70 + 0.47)
| Location | 16,698,753 – 16,698,882 |
|---|---|
| Length | 129 |
| Sequences | 13 |
| Columns | 148 |
| Reading direction | reverse |
| Mean pairwise identity | 60.06 |
| Shannon entropy | 0.83426 |
| G+C content | 0.31766 |
| Mean single sequence MFE | -21.79 |
| Consensus MFE | -7.72 |
| Energy contribution | -8.44 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.65 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.49 |
| SVM RNA-class probability | 0.998785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
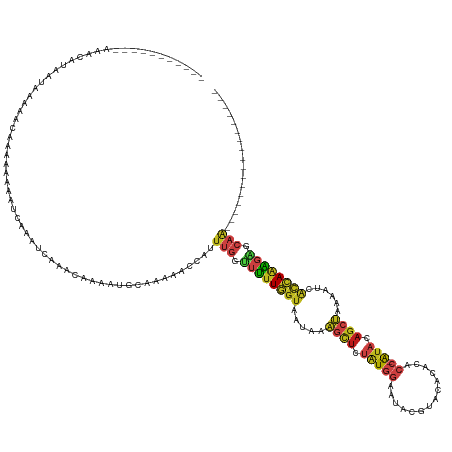
>dm3.chr2L 16698753 129 - 23011544 -UAACGCGAGAAAAGUAAAGAAUACCAAAUAAAAUCAACUCAAACAUAAUGCAGAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAAUAUACAUAAACACC-AUACAGCUAAAAUCACCAAAGAGCAA----------------- -...(....)............................(((..........((((((((....)))))))).......((((.(((((..............))-))).))))............)))...----------------- ( -20.84, z-score = -2.59, R) >droSim1.chrX 10558594 143 - 17042790 -UAACAGA--AAAUUACAUUACAAA-AUUCAACAACGAAAACUUUUGGCAUUACAAAUUAAUGCAUUUUUAAAUACAAUAUUUAUUGAUAUACCUAGAUUUAAA-GCGCAGCAACAACAACCAGGCGAAAGCGAUGAAAAAAUUCCAA -.......--......((((.((((-((((......)))...)))))((((((.....))))))..((((((((...(((((....)))))......)))))))-)..................((....))))))............ ( -15.40, z-score = -0.62, R) >droSec1.super_7 345765 127 - 3727775 -UAACACUAGAAAAGUAAAUUAUACCAAAUAAAAUCAACUCAAGCAUAAUGCAGAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAAUACUUAUA--CACC-AUACAGCUAAAAUCACCAAAGACCAA----------------- -..........................................((.....)).........((((((((((((.((..((((.(((((..........--..))-))).))))...)).))))))))))))----------------- ( -24.80, z-score = -3.98, R) >droYak2.chr2R 16734897 119 + 21139217 -----------UAAAGUUAGAAAAUGAAAAAAAAUCAACUCAAGCAUAAUGCAGAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAACACAUAAACACACC-AUACAGCUAAAAUCACCAAAGAGCAA----------------- -----------....(((.((...(((.......)))..)).)))....((((((((((....)))))))(((.((..((((.(((((..............))-))).))))...)).))).....))).----------------- ( -21.64, z-score = -2.79, R) >droEre2.scaffold_4845 19768399 128 - 22589142 UAACGUUAGAAAAAUGUAACAAAACGAAAAAAAAUCAACUCAAGCAUAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAACACAUA--CACACC-AUACAGCUAAAAUCACCAAAGAGCAA----------------- ....((((.(....).))))..................(((..((.....))(((((((....)))))))(((.((..((((.(((((........--....))-))).))))...)).)))...)))...----------------- ( -22.10, z-score = -2.87, R) >droAna3.scaffold_12916 316217 117 + 16180835 ------UAAAAAAUCAUAACCACAACAAAAGCAACUCA---GAAA-AAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAAUAUG---AGACACC-AUACAGCUAAAAUCACCAAAGAACAA----------------- ------.......((...(((.........(((.....---....-...)))(((((((....)))))))))).....((((.(((((......---.....))-))).))))............))....----------------- ( -18.90, z-score = -2.62, R) >dp4.chr4_group3 8444203 118 - 11692001 -----------AAACAUUUUAAAAACAUAAGCAACUCAAGCGAAA-AAAUGCAAAAACCAUUUGGGUUUUGGUGAUAAAGCUCUAUGGAUCUUUGAUAUACACC-AUACAGCUAAAAUCACCAAAGAUCAA----------------- -----------............................(((...-...))).........(((..((((((((((..((((.(((((..............))-))).))))...))))))))))..)))----------------- ( -23.74, z-score = -2.80, R) >droPer1.super_1 9895322 118 - 10282868 -----------AAACAUUUUAAAAACAUAAGCAACUCAAGCGAAA-AAAUGCAAAAACCAUUUGGGUUUUGGUGAUAAAGCUCUAUGGAUCUUUGAUAUACACC-AUACAGCUAAAAUCACCAAAGAUCAA----------------- -----------............................(((...-...))).........(((..((((((((((..((((.(((((..............))-))).))))...))))))))))..)))----------------- ( -23.74, z-score = -2.80, R) >droWil1.scaffold_181132 161966 130 + 1035393 AAAUGUAAUUAAAUCAAAAUCAAUACGAAAGAAAACAACUCAAGCAAAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAUUAUGCUCAUACACC-AUACAGCUAAAAUCACCAAAGACCAA----------------- (((((....................(....)............((.....))......)))))((((((((((.((..((((.(((((..(((....)))..))-))).))))...)).))))))))))..----------------- ( -27.70, z-score = -4.61, R) >droVir3.scaffold_12963 7343316 103 + 20206255 -----------------------CAAGUGAACAA--UAAAAGAAAGCAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAAUACG--CGCACACC-AUACAGCUAAAAUCACCAAAGAGCAA----------------- -----------------------...((((....--...............((((((((....)))))))).......((((.(((((......--......))-))).))))....))))..........----------------- ( -21.20, z-score = -2.17, R) >droMoj3.scaffold_6500 7202316 105 - 32352404 -----------------------UAACUGAAUAAACGAAAAGAAAGCAAUGCAAAGACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAAUACG--CGCACACC-AUACAGCUAAAAUCACCAAAGAACAA----------------- -----------------------....(((.....................((((((((....)))))))).......((((.(((((......--......))-))).))))....)))...........----------------- ( -18.20, z-score = -1.70, R) >droGri2.scaffold_14978 831307 104 - 1124632 -----------------------UAACGAAGGAAAAAAAGAAAGC-AAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUGUGGAUUAUG--CGCACACC-AUAGAGCUAAAAUCACCAAAGAGCAA----------------- -----------------------...(....)...........((-....)).........(((.((((((((.((..((((((((((......--......))-))))))))...)).)))))))).)))----------------- ( -26.60, z-score = -3.48, R) >anoGam1.chr3R 47383922 127 + 53272125 -UGAUAAAUUCAAUUUUGUGGUGAACAUUCAAUGUAAGCUAAGUUAAACGAAAGAAACACAACGACUGGUAGUAAUAAAAACUCGUGGAAUACAUGCACAAAAAGGUAUAGAGUAAAGAGUUAAUCGA-------------------- -.(((.(((((.((((((((.(.........(((((..((........(....).......((((.(..((....))...).))))))..))))).........).))))))))...))))).)))..-------------------- ( -18.47, z-score = 0.32, R) >consensus ___________AAACAUAAUAAAAACAAAAAAAAUCAAAUCAAACAAAAUGCAAAAACCAUUUGGUUUUUGGUAAUAAAGCUCUAUGGAAUACGUACACACACC_AUACAGCUAAAAUCACCAAAGAGCAA_________________ .............................................................(((.((((((((.....((((.(((((..............)).))).))))......)))))))).)))................. ( -7.72 = -8.44 + 0.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:10 2011