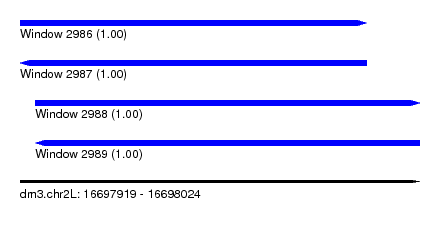
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,697,919 – 16,698,024 |
| Length | 105 |
| Max. P | 0.999362 |

| Location | 16,697,919 – 16,698,010 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 84.13 |
| Shannon entropy | 0.32911 |
| G+C content | 0.33717 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -24.05 |
| Energy contribution | -24.23 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.08 |
| Mean z-score | -5.32 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.59 |
| SVM RNA-class probability | 0.999003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16697919 91 + 23011544 -----------------UAUCAUUUUU-GCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCACGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCU -----------------..........-((((...(((((((((...((((.(((((.((...(((.....)))..)).))))).))))...)))))))))...)))). ( -26.40, z-score = -5.10, R) >droSec1.super_7 344902 91 + 3727775 -----------------UAUCAUUUUU-GCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCACGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCU -----------------..........-((((...(((((((((...((((.(((((.((...(((.....)))..)).))))).))))...)))))))))...)))). ( -26.40, z-score = -5.10, R) >droYak2.chr2R 16734045 91 - 21139217 -----------------UAUCAUUUUU-GCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCGCGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCU -----------------..........-((((...(((((((((...((((.(((((.((...(((.....)))..)).))))).))))...)))))))))...)))). ( -26.70, z-score = -4.86, R) >droEre2.scaffold_4845 19767530 91 + 22589142 -----------------UAUCAUUUUU-GCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCGCGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCU -----------------..........-((((...(((((((((...((((.(((((.((...(((.....)))..)).))))).))))...)))))))))...)))). ( -26.70, z-score = -4.86, R) >droAna3.scaffold_12916 315266 91 - 16180835 -----------------CAUUAUUUUU-GCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCACGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCU -----------------..........-((((...(((((((((...((((.(((((.((...(((.....)))..)).))))).))))...)))))))))...)))). ( -26.40, z-score = -5.06, R) >dp4.chr4_group3 8443182 99 + 11692001 ---------UGUUCUCCUUUAAUUUUU-GCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUUUUGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCU ---------..................-((((...(((((((((...((((.(((((.((....(((.....))).)).))))).))))...)))))))))...)))). ( -27.60, z-score = -5.40, R) >droPer1.super_1 9894304 99 + 10282868 ---------UGUUCUCCUUUAAUUUUU-GCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUUUUGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCU ---------..................-((((...(((((((((...((((.(((((.((....(((.....))).)).))))).))))...)))))))))...)))). ( -27.60, z-score = -5.40, R) >droWil1.scaffold_181132 160059 109 - 1035393 UUUUCCUCUCCCUCCAUCAUCAUUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUUUUGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCU ............................((((...(((((((((...((((.(((((.((....(((.....))).)).))))).))))...)))))))))...)))). ( -27.60, z-score = -6.23, R) >droVir3.scaffold_12963 7342314 88 - 20206255 -----------------UUUCAUUUUU-GCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUUU---ACAGUUUGUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCU -----------------..........-((((...(((((((((...((((.(((((.(....(---((.....))).)))))).))))...)))))))))...)))). ( -26.90, z-score = -5.90, R) >droMoj3.scaffold_6500 7201340 88 + 32352404 -----------------UUUCAUUUUU-GCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUUC---ACAUAUUGUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCU -----------------..........-((((...(((((((((...((((.(((((.((...(---(.....)).)).))))).))))...)))))))))...)))). ( -25.90, z-score = -5.82, R) >droGri2.scaffold_14978 830202 95 + 1124632 ----------CAAUUUCUUUCAUUUUU-GCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCU---AUAUCUUGUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCU ----------.................-((((...(((((((((...((((.(((((.(...(.---.......)...)))))).))))...)))))))))...)))). ( -25.60, z-score = -5.74, R) >anoGam1.chr3R 47380871 83 - 53272125 ------------------------UCCGGCUGUAUCUUUGGUAUUCUAGCUGUAGAAUGUUGUUU--UCAUUGUAAUAUCUCUAAAGCUUUAGUACCAGAGGUCCAACU ------------------------...((.((.((((((((((((..((((.((((((((((...--......)))))).)))).))))..)))))))))))).)).)) ( -26.20, z-score = -4.40, R) >consensus _________________UAUCAUUUUU_GCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCUCGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCU ............................((((...(((((((((...((((.(((((.((................)).))))).))))...)))))))))...)))). (-24.05 = -24.23 + 0.18)

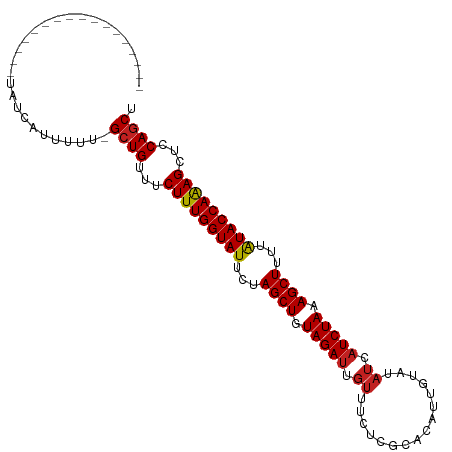
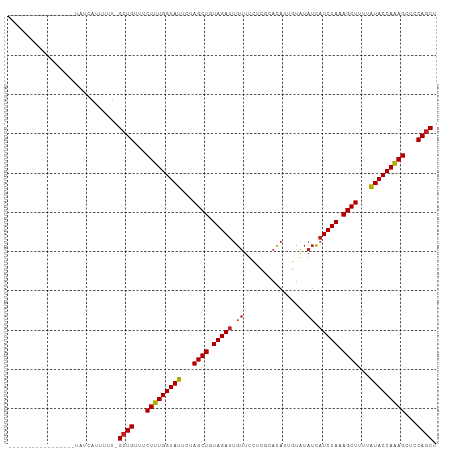
| Location | 16,697,919 – 16,698,010 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 84.13 |
| Shannon entropy | 0.32911 |
| G+C content | 0.33717 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -24.90 |
| Energy contribution | -24.83 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -5.20 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.50 |
| SVM RNA-class probability | 0.998804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16697919 91 - 23011544 AGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCGUGAAACAAUCUACAGCUAGAAUACCAAAGAAACAGC-AAAAAUGAUA----------------- .((((...(((((((((...((((.(((((....(((.......))).....))))).))))...)))))))))...))))-..........----------------- ( -28.60, z-score = -5.46, R) >droSec1.super_7 344902 91 - 3727775 AGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCGUGAAACAAUCUACAGCUAGAAUACCAAAGAAACAGC-AAAAAUGAUA----------------- .((((...(((((((((...((((.(((((....(((.......))).....))))).))))...)))))))))...))))-..........----------------- ( -28.60, z-score = -5.46, R) >droYak2.chr2R 16734045 91 + 21139217 AGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCGCGAAACAAUCUACAGCUAGAAUACCAAAGAAACAGC-AAAAAUGAUA----------------- .((((...(((((((((...((((.(((((......................))))).))))...)))))))))...))))-..........----------------- ( -27.05, z-score = -4.82, R) >droEre2.scaffold_4845 19767530 91 - 22589142 AGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCGCGAAACAAUCUACAGCUAGAAUACCAAAGAAACAGC-AAAAAUGAUA----------------- .((((...(((((((((...((((.(((((......................))))).))))...)))))))))...))))-..........----------------- ( -27.05, z-score = -4.82, R) >droAna3.scaffold_12916 315266 91 + 16180835 AGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCGUGAAACAAUCUACAGCUAGAAUACCAAAGAAACAGC-AAAAAUAAUG----------------- .((((...(((((((((...((((.(((((....(((.......))).....))))).))))...)))))))))...))))-..........----------------- ( -28.60, z-score = -5.65, R) >dp4.chr4_group3 8443182 99 - 11692001 AGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCAAAAAACAAUCUACAGCUAGAAUACCAAAGAAACAGC-AAAAAUUAAAGGAGAACA--------- .((((...(((((((((...((((.(((((......................))))).))))...)))))))))...))))-..................--------- ( -27.05, z-score = -5.16, R) >droPer1.super_1 9894304 99 - 10282868 AGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCAAAAAACAAUCUACAGCUAGAAUACCAAAGAAACAGC-AAAAAUUAAAGGAGAACA--------- .((((...(((((((((...((((.(((((......................))))).))))...)))))))))...))))-..................--------- ( -27.05, z-score = -5.16, R) >droWil1.scaffold_181132 160059 109 + 1035393 AGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCAAAAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAAUGAUGAUGGAGGGAGAGGAAAA .((((...(((((((((...((((.(((((......................))))).))))...)))))))))...))))............................ ( -27.05, z-score = -4.32, R) >droVir3.scaffold_12963 7342314 88 + 20206255 AGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUACAAACUGU---AAAAACAAUCUACAGCUAGAAUACCAAAGAAACAGC-AAAAAUGAAA----------------- .((((...(((((((((...((((.(((((..(((.....))---)......))))).))))...)))))))))...))))-..........----------------- ( -28.10, z-score = -6.68, R) >droMoj3.scaffold_6500 7201340 88 - 32352404 AGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUACAAUAUGU---GAAAACAAUCUACAGCUAGAAUACCAAAGAAACAGC-AAAAAUGAAA----------------- .((((...(((((((((...((((.(((((.........(((---....)))))))).))))...)))))))))...))))-..........----------------- ( -28.80, z-score = -6.85, R) >droGri2.scaffold_14978 830202 95 - 1124632 AGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUACAAGAUAU---AGAAACAAUCUACAGCUAGAAUACCAAAGAAACAGC-AAAAAUGAAAGAAAUUG---------- .((((...(((((((((...((((.(((((............---.......))))).))))...)))))))))...))))-.................---------- ( -27.21, z-score = -6.40, R) >anoGam1.chr3R 47380871 83 + 53272125 AGUUGGACCUCUGGUACUAAAGCUUUAGAGAUAUUACAAUGA--AAACAACAUUCUACAGCUAGAAUACCAAAGAUACAGCCGGA------------------------ .((((...((.(((((.(..((((.(((((............--........))))).))))..).))))).))...))))....------------------------ ( -16.05, z-score = -1.59, R) >consensus AGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCGAAAAACAAUCUACAGCUAGAAUACCAAAGAAACAGC_AAAAAUGAUA_________________ .((((...(((((((((...((((.(((((......................))))).))))...)))))))))...))))............................ (-24.90 = -24.83 + -0.06)
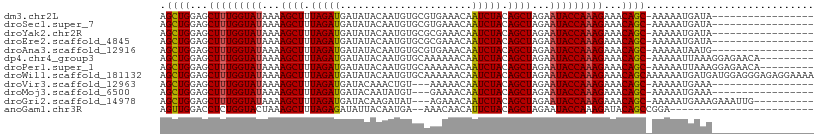
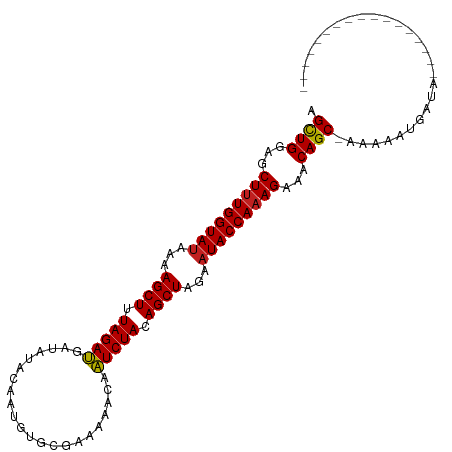
| Location | 16,697,923 – 16,698,024 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 87.14 |
| Shannon entropy | 0.26556 |
| G+C content | 0.32839 |
| Mean single sequence MFE | -28.21 |
| Consensus MFE | -24.05 |
| Energy contribution | -24.23 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.08 |
| Mean z-score | -4.51 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.82 |
| SVM RNA-class probability | 0.999362 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
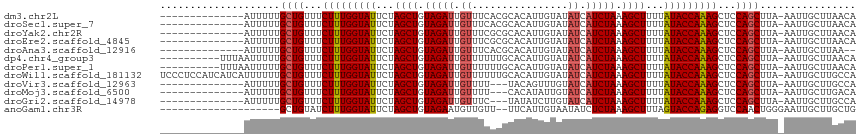
>dm3.chr2L 16697923 101 + 23011544 --------------AUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCACGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCUUA-AAUUGCUUAACA --------------((((..((((...(((((((((...((((.(((((.((...(((.....)))..)).))))).))))...)))))))))...))))..)-)))......... ( -27.40, z-score = -4.44, R) >droSec1.super_7 344906 101 + 3727775 --------------AUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCACGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCUUA-AAUUGCUUAACA --------------((((..((((...(((((((((...((((.(((((.((...(((.....)))..)).))))).))))...)))))))))...))))..)-)))......... ( -27.40, z-score = -4.44, R) >droYak2.chr2R 16734049 101 - 21139217 --------------AUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCGCGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCUUA-AAUUGCUUAACA --------------((((..((((...(((((((((...((((.(((((.((...(((.....)))..)).))))).))))...)))))))))...))))..)-)))......... ( -27.70, z-score = -4.19, R) >droEre2.scaffold_4845 19767534 101 + 22589142 --------------AUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCGCGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCUUA-AAUUGCUUAACA --------------((((..((((...(((((((((...((((.(((((.((...(((.....)))..)).))))).))))...)))))))))...))))..)-)))......... ( -27.70, z-score = -4.19, R) >droAna3.scaffold_12916 315270 99 - 16180835 --------------AUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCACGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCUUA-AAUUGCUUAA-- --------------((((..((((...(((((((((...((((.(((((.((...(((.....)))..)).))))).))))...)))))))))...))))..)-))).......-- ( -27.40, z-score = -4.61, R) >dp4.chr4_group3 8443190 105 + 11692001 ----------UUUAAUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUUUUGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCUUA-AAUUGCUUAACA ----------..((((((..((((...(((((((((...((((.(((((.((....(((.....))).)).))))).))))...)))))))))...))))..)-)))))....... ( -31.10, z-score = -5.28, R) >droPer1.super_1 9894312 105 + 10282868 ----------UUUAAUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUUUUGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCUUA-AAUUGCUUAACA ----------..((((((..((((...(((((((((...((((.(((((.((....(((.....))).)).))))).))))...)))))))))...))))..)-)))))....... ( -31.10, z-score = -5.28, R) >droWil1.scaffold_181132 160067 115 - 1035393 UCCCUCCAUCAUCAUUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUUUUGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCUUA-AAUUGCUUGCCA ............((.(((..((((...(((((((((...((((.(((((.((....(((.....))).)).))))).))))...)))))))))...))))..)-)).))....... ( -29.30, z-score = -4.82, R) >droVir3.scaffold_12963 7342318 98 - 20206255 --------------AUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUU---UACAGUUUGUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCUUA-AAUUGCUUGCCA --------------((((..((((...(((((((((...((((.(((((.(....---(((.....))).)))))).))))...)))))))))...))))..)-)))......... ( -27.90, z-score = -4.61, R) >droMoj3.scaffold_6500 7201344 98 + 32352404 --------------AUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUU---CACAUAUUGUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCUUA-AAUUGCUUGACA --------------((((..((((...(((((((((...((((.(((((.((...---((.....)).)).))))).))))...)))))))))...))))..)-)))......... ( -26.90, z-score = -4.40, R) >droGri2.scaffold_14978 830213 98 + 1124632 --------------AUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUC---UAUAUCUUGUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCUUA-AAUUGCUUGCCA --------------((((..((((...(((((((((...((((.(((((.(...(---........)...)))))).))))...)))))))))...))))..)-)))......... ( -26.60, z-score = -4.76, R) >anoGam1.chr3R 47380875 94 - 53272125 --------------------GCUGUAUCUUUGGUAUUCUAGCUGUAGAAUGUUGUU--UUCAUUGUAAUAUCUCUAAAGCUUUAGUACCAGAGGUCCAACUGGGAAUUGCUUGCUG --------------------((.((((((((((((((..((((.((((((((((..--.......)))))).)))).))))..)))))))))))(((.....)))..)))..)).. ( -28.00, z-score = -3.15, R) >consensus ______________AUUUUUGCUGUUUCUUUGGUAUUCUAGCUGUAGAUUGUUUCUCGCACAUUGUAUAUCAUCUAAAGCUUUUAUACCAAAGCUCCAGCUUA_AAUUGCUUAACA ....................((((...(((((((((...((((.(((((.((................)).))))).))))...)))))))))...))))................ (-24.05 = -24.23 + 0.18)

| Location | 16,697,923 – 16,698,024 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 87.14 |
| Shannon entropy | 0.26556 |
| G+C content | 0.32839 |
| Mean single sequence MFE | -26.77 |
| Consensus MFE | -26.25 |
| Energy contribution | -26.18 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.98 |
| Structure conservation index | 0.98 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999011 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 16697923 101 - 23011544 UGUUAAGCAAUU-UAAGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCGUGAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAU-------------- ............-...((((...(((((((((...((((.(((((....(((.......))).....))))).))))...)))))))))...))))......-------------- ( -28.60, z-score = -4.35, R) >droSec1.super_7 344906 101 - 3727775 UGUUAAGCAAUU-UAAGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCGUGAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAU-------------- ............-...((((...(((((((((...((((.(((((....(((.......))).....))))).))))...)))))))))...))))......-------------- ( -28.60, z-score = -4.35, R) >droYak2.chr2R 16734049 101 + 21139217 UGUUAAGCAAUU-UAAGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCGCGAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAU-------------- ............-...((((...(((((((((...((((.(((((......................))))).))))...)))))))))...))))......-------------- ( -27.05, z-score = -3.73, R) >droEre2.scaffold_4845 19767534 101 - 22589142 UGUUAAGCAAUU-UAAGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCGCGAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAU-------------- ............-...((((...(((((((((...((((.(((((......................))))).))))...)))))))))...))))......-------------- ( -27.05, z-score = -3.73, R) >droAna3.scaffold_12916 315270 99 + 16180835 --UUAAGCAAUU-UAAGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCGUGAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAU-------------- --..........-...((((...(((((((((...((((.(((((....(((.......))).....))))).))))...)))))))))...))))......-------------- ( -28.60, z-score = -4.88, R) >dp4.chr4_group3 8443190 105 - 11692001 UGUUAAGCAAUU-UAAGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCAAAAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAUUAAA---------- ........((((-(..((((...(((((((((...((((.(((((......................))))).))))...)))))))))...))))..)))))...---------- ( -27.45, z-score = -3.91, R) >droPer1.super_1 9894312 105 - 10282868 UGUUAAGCAAUU-UAAGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCAAAAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAUUAAA---------- ........((((-(..((((...(((((((((...((((.(((((......................))))).))))...)))))))))...))))..)))))...---------- ( -27.45, z-score = -3.91, R) >droWil1.scaffold_181132 160067 115 + 1035393 UGGCAAGCAAUU-UAAGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCAAAAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAAUGAUGAUGGAGGGA ............-...((((...(((((((((...((((.(((((......................))))).))))...)))))))))...)))).................... ( -27.05, z-score = -2.80, R) >droVir3.scaffold_12963 7342318 98 + 20206255 UGGCAAGCAAUU-UAAGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUACAAACUGUA---AAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAU-------------- ............-...((((...(((((((((...((((.(((((..(((.....)))---......))))).))))...)))))))))...))))......-------------- ( -28.10, z-score = -4.97, R) >droMoj3.scaffold_6500 7201344 98 - 32352404 UGUCAAGCAAUU-UAAGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUACAAUAUGUG---AAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAU-------------- ............-...((((...(((((((((...((((.(((((.........(((.---...)))))))).))))...)))))))))...))))......-------------- ( -28.80, z-score = -5.27, R) >droGri2.scaffold_14978 830213 98 - 1124632 UGGCAAGCAAUU-UAAGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUACAAGAUAUA---GAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAU-------------- ............-...((((...(((((((((...((((.(((((.............---......))))).))))...)))))))))...))))......-------------- ( -27.21, z-score = -5.17, R) >anoGam1.chr3R 47380875 94 + 53272125 CAGCAAGCAAUUCCCAGUUGGACCUCUGGUACUAAAGCUUUAGAGAUAUUACAAUGAA--AACAACAUUCUACAGCUAGAAUACCAAAGAUACAGC-------------------- ................((((...((.(((((.(..((((.(((((.............--.......))))).))))..).))))).))...))))-------------------- ( -15.25, z-score = -0.71, R) >consensus UGUUAAGCAAUU_UAAGCUGGAGCUUUGGUAUAAAAGCUUUAGAUGAUAUACAAUGUGCGAAAAACAAUCUACAGCUAGAAUACCAAAGAAACAGCAAAAAU______________ ................((((...(((((((((...((((.(((((......................))))).))))...)))))))))...)))).................... (-26.25 = -26.18 + -0.06)
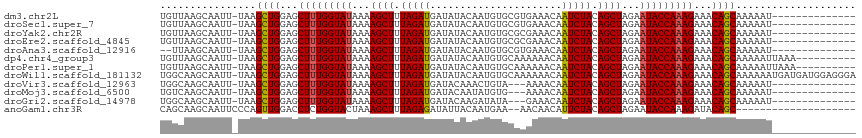

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:07 2011