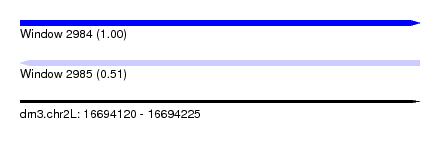
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,694,120 – 16,694,225 |
| Length | 105 |
| Max. P | 0.995527 |

| Location | 16,694,120 – 16,694,225 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 61.41 |
| Shannon entropy | 0.71071 |
| G+C content | 0.53307 |
| Mean single sequence MFE | -31.22 |
| Consensus MFE | -14.62 |
| Energy contribution | -15.52 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.995527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
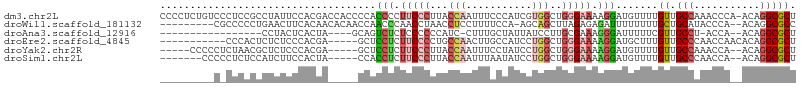
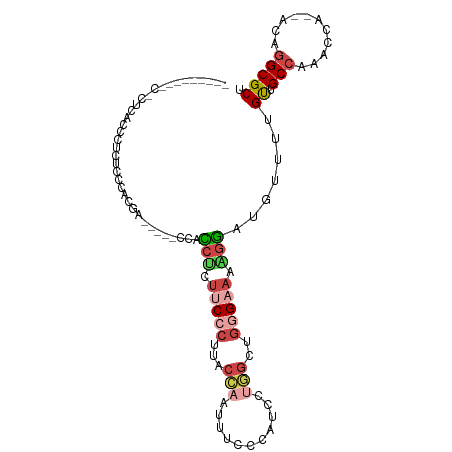
>dm3.chr2L 16694120 105 + 23011544 AGCGCCUGU-UGGGUUUGGCAACAAAACAUCCUUUUCCCAGCCACGAUGGGAAAUUGGUAAGGGAAGGGGUGGGGUGGUCGUGGAAUAGGCGGAGGGACAGAGGGG ..(((((((-(.....((((.((....(((((((((((..((((...........))))..)))))))))))..)).))))...)))))))).............. ( -35.30, z-score = -1.47, R) >droWil1.scaffold_181132 155375 94 - 1035393 GGCGCCUGU--UGGGUAUGCAGCAAAAAAAAUCUCUCUAAGCUGCU-UGGAAAAGGAGGUUAGGUUGGGUUGGUUGUGUUGUGAAGUUCAGGGGGCG--------- ..(((((.(--((..(..((((((.((..(((((.(((((.((.((-(....))).)).)))))..)))))..)).))))))...)..))).)))))--------- ( -34.80, z-score = -4.29, R) >droAna3.scaffold_12916 311354 81 - 16180835 AGCGCCUGU--UGGU-AGGCAACGAAAAAUCCCUUUCGCAAGGAUAAUAGCAAAG-GAUGGGGGGAGAGACUGC----UAGUGAGUAGG----------------- ...(((((.--...)-)))).........((((((((....)))......((...-..)).)))))....((((----(....))))).----------------- ( -19.20, z-score = 0.23, R) >droEre2.scaffold_4845 19763746 90 + 22589142 AGCGCCUGUGUUGGUUGGGCAACAAAGCAUCCUUUUCCCAGCCAGGAUGGCAAGUUGGCAGGGGAAGAGGAGC-----UCGUGGGAGAGAGUGGG----------- ...((...(((((......)))))..)).((((((((((.(((((.........)))))..))))))))))((-----((........))))...----------- ( -32.80, z-score = -0.92, R) >droYak2.chr2R 16730174 94 - 21139217 AGCGCCUGU--UGGUUUGGCAACAAAACAUCCUUUUCCCAGCCAGGAUAGGAAAUUGGUAAGGGAAGAGGAGC-----UCGUGGGAGAGCGUUAGAGGGGG----- ..(.(((..--((.((((....)))).))((((((((((.(((((.........)))))..))))))))))((-----((......)))).....))).).----- ( -35.30, z-score = -2.72, R) >droSim1.chr2L 16390287 92 + 22036055 AGCGCCUGU--UGGUUGGGCAACAAAACAUCCUUUUCCCAGCCAGGAUAUUAAAUUGGUAAGGGAAGAGGUGG-----UAGUGGAAGAUGGAGAGGGGG------- ..(.(((.(--(.(((......((..((..(((((((((.(((((.........)))))..)))))))))..)-----)..))...))).)).))).).------- ( -29.90, z-score = -2.14, R) >consensus AGCGCCUGU__UGGUUAGGCAACAAAACAUCCUUUUCCCAGCCAGGAUAGCAAAUUGGUAAGGGAAGAGGUGG_____UCGUGGAAGAGAGGGAG_G_________ .(((((((.......)))))..........(((((((((.(((((.........)))))..)))))))))..........))........................ (-14.62 = -15.52 + 0.90)


| Location | 16,694,120 – 16,694,225 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 61.41 |
| Shannon entropy | 0.71071 |
| G+C content | 0.53307 |
| Mean single sequence MFE | -19.63 |
| Consensus MFE | -6.56 |
| Energy contribution | -7.70 |
| Covariance contribution | 1.14 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.26 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.513856 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16694120 105 - 23011544 CCCCUCUGUCCCUCCGCCUAUUCCACGACCACCCCACCCCUUCCCUUACCAAUUUCCCAUCGUGGCUGGGAAAAGGAUGUUUUGUUGCCAAACCCA-ACAGGCGCU ..............(((((.................((..(((((...(((...........)))..)))))..)).......((((.......))-))))))).. ( -20.00, z-score = -0.12, R) >droWil1.scaffold_181132 155375 94 + 1035393 ---------CGCCCCCUGAACUUCACAACACAACCAACCCAACCUAACCUCCUUUUCCA-AGCAGCUUAGAGAGAUUUUUUUUGCUGCAUACCCA--ACAGGCGCC ---------((((...((.....))..................................-.(((((..(((((...)))))..))))).......--...)))).. ( -14.80, z-score = -2.16, R) >droAna3.scaffold_12916 311354 81 + 16180835 -----------------CCUACUCACUA----GCAGUCUCUCCCCCCAUC-CUUUGCUAUUAUCCUUGCGAAAGGGAUUUUUCGUUGCCU-ACCA--ACAGGCGCU -----------------..........(----((.............(((-((((((..........))))..)))))........((((-....--..))))))) ( -14.60, z-score = -0.46, R) >droEre2.scaffold_4845 19763746 90 - 22589142 -----------CCCACUCUCUCCCACGA-----GCUCCUCUUCCCCUGCCAACUUGCCAUCCUGGCUGGGAAAAGGAUGCUUUGUUGCCCAACCAACACAGGCGCU -----------................(-----((((((.(((((..((((...........)))).))))).)))).(((((((((......))))).))))))) ( -27.40, z-score = -2.32, R) >droYak2.chr2R 16730174 94 + 21139217 -----CCCCCUCUAACGCUCUCCCACGA-----GCUCCUCUUCCCUUACCAAUUUCCUAUCCUGGCUGGGAAAAGGAUGUUUUGUUGCCAAACCA--ACAGGCGCU -----..........((((.......((-----((((((.(((((...(((...........)))..))))).)))).))))(((((......))--))))))).. ( -23.50, z-score = -1.83, R) >droSim1.chr2L 16390287 92 - 22036055 -------CCCCCUCUCCAUCUUCCACUA-----CCACCUCUUCCCUUACCAAUUUAAUAUCCUGGCUGGGAAAAGGAUGUUUUGUUGCCCAACCA--ACAGGCGCU -------.....................-----((.(((.(((((...(((...........)))..))))).)))......(((((......))--))))).... ( -17.50, z-score = -0.65, R) >consensus _________C_CUCACCCUCUCCCACGA_____CCACCUCUUCCCUUACCAAUUUCCCAUCCUGGCUGGGAAAAGGAUGUUUUGUUGCCAAACCA__ACAGGCGCU ....................................(((.(((((...(((...........)))..))))).))).......((.(((...........))))). ( -6.56 = -7.70 + 1.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:04 2011