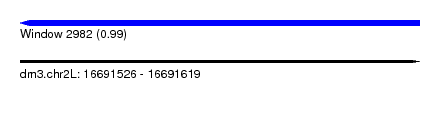
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,691,526 – 16,691,619 |
| Length | 93 |
| Max. P | 0.990518 |

| Location | 16,691,526 – 16,691,619 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 70.62 |
| Shannon entropy | 0.56351 |
| G+C content | 0.45158 |
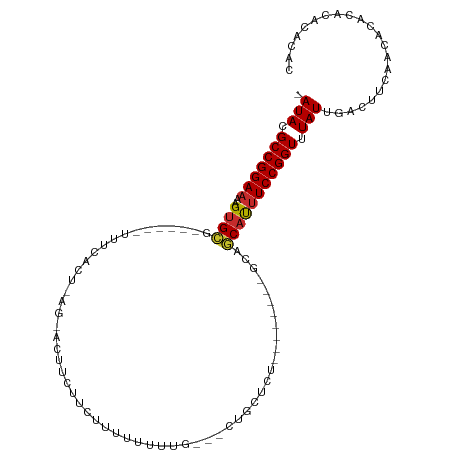
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -10.59 |
| Energy contribution | -10.65 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.42 |
| SVM RNA-class probability | 0.990518 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
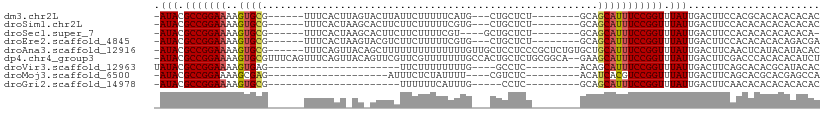
>dm3.chr2L 16691526 93 - 23011544 -AUACGCCGGAAAAGUGCG------UUUCACUUAGUACUUAUUCUUUUUCAUG---CUGCUCU--------GCAGCAUUUCCGGUUUAUUGACUUCCACGCACACACACAC -(((.(((((((((((((.------.........))))))..........(((---((((...--------)))))))))))))).)))...................... ( -24.30, z-score = -3.68, R) >droSim1.chr2L 16387702 93 - 22036055 -AUACGCCGGAAAAGUGCG------UUUCACUAAGCACUUCUUCUUUUUCGUG---CUGCUCU--------GCAGCAUUUCCGGUUUAUUGACUUCCACACACACACACAC -(((.(((((((((((((.------.........))))))..........(((---((((...--------)))))))))))))).)))...................... ( -26.00, z-score = -4.19, R) >droSec1.super_7 338493 91 - 3727775 -AUACGCCGGAAAAGUGCG------UUUCACUAAGCACUUCUUCUUUUCGU----GCUGCUCU--------GCAGCAUUUCCGGUUUAUUGACUUCCACACACACACACA- -(((.(((((((((((((.------.........)))))).........((----(((((...--------)))))))))))))).))).....................- ( -26.00, z-score = -4.07, R) >droEre2.scaffold_4845 19761248 93 - 22589142 -AUACGCCGGAAAAGUGCG------UUUCACUAAGUACGUCUUCUUUUUCGUG---CUGCUCU--------GCAGCAUUUCCGGUUUAUUGACUUCCACACACACAGACGA -(((.((((((((.((((.------.........)))).............((---((((...--------)))))))))))))).)))...................... ( -23.00, z-score = -1.52, R) >droAna3.scaffold_12916 309251 104 + 16180835 -AUACGCCGGAAAAGUGCG------UUUCAGUUACAGCUUUUUUUUUUUUUUGUUGCUCCUCCCGCUCUGUGCUGCAUUUCCGGUUUAUUGACUUCAACUCAUACAUACAC -(((.(((((((..((((.------.........((((..............))))........((.....)).))))))))))).)))...................... ( -17.84, z-score = -0.69, R) >dp4.chr4_group3 8434504 108 - 11692001 -AUACGCCGGAAAAGUGCGUUUCAGUUUCAGUUACAGUUCGUUCGUUUUUUUGCCACUGCUCUGCGGCA--GAAGCAUUUCCGGUUUAUUGACUUCGACCCACACACAUCU -(((.(((((((..((((((..(.......)..)).............(((((((.(......).))))--)))))))))))))).)))...................... ( -24.10, z-score = -0.58, R) >droVir3.scaffold_12963 7340454 76 + 20206255 UAUACGCCGGAAAAGUGAG----------------------UUCUUUUUUUUG----GCCUC---------ACAGCAUUUCCGGUUUAUUGACUUCAGCACACGCAUACAC .(((.((((((((.(((((----------------------..(........)----..)))---------))....)))))))).)))........((....))...... ( -20.10, z-score = -3.27, R) >droMoj3.scaffold_6500 7199603 77 - 32352404 -AUACGCCGGAAAAGCGAG--------------------AUUUCUCUAUUUU----CGUCUC---------ACAUCACGUCCGGUUUAUUGACUUCAGCACGCACGAGCCA -(((.((((((...(.(((--------------------((...........----.)))))---------.)......)))))).)))............((....)).. ( -13.20, z-score = 0.41, R) >droGri2.scaffold_14978 828728 74 - 1124632 -AUACGCCGGAAAAGUGCG----------------------UUUUUUCAUUUG-----CCUC---------GCAGCAUUUCCGGUUUAUUGACUUCAACACACACACACAC -(((.((((((((.(((((----------------------............-----...)---------))).).)))))))).)))...................... ( -15.76, z-score = -2.19, R) >consensus _AUACGCCGGAAAAGUGCG______UUUCACU_AG_ACUUCUUCUUUUUUUUG___CUGCUCU________GCAGCAUUUCCGGUUUAUUGACUUCAACACACACACACAC .(((.(((((((..((((........................................................))))))))))).)))...................... (-10.59 = -10.65 + 0.06)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:01 2011