
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,689,120 – 16,689,231 |
| Length | 111 |
| Max. P | 0.765569 |

| Location | 16,689,120 – 16,689,231 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 94.95 |
| Shannon entropy | 0.08904 |
| G+C content | 0.49730 |
| Mean single sequence MFE | -21.27 |
| Consensus MFE | -19.32 |
| Energy contribution | -19.52 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.695027 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16689120 111 + 23011544 GUCCUCCAUUUCCCUUUGCCUACGUUUCCUCCACUCUUUCCAUUAGGCUGGCCAUCACAUUUGAGGUUUUAUCCUUGGGCUGUCAACACGUGCUUUGACACGCCCCCUUUA ............((...(((((.....................))))).))...........((((......))))((((((((((........)))))).))))...... ( -22.30, z-score = -1.42, R) >droEre2.scaffold_4845 19758985 111 + 22589142 GUCCUGCAUUUCCCAUCGCCUACGUUUCCUCCUCUCUUCCCAUUAGGCUGCCCAUCACUUUUGAGGUUUUAUCCUUGGGCUGUCAACACGUGCUUUGACACGCCCCCUUUA .................(((((.....................)))))..............((((......))))((((((((((........)))))).))))...... ( -21.10, z-score = -1.38, R) >droYak2.chr2R 16725363 111 - 21139217 GUCCUCCAUUUUCCUUUGCCUACGUUUCCUCCUCUCCUUCCAUUAGGCUGCCCAUCACUUUUGAGGUUUUAUCCUUGGGCUGUCAACACGUGCUUUGACACGCCCCUUUAA .................(((((.....................)))))..............((((......))))((((((((((........)))))).))))...... ( -20.80, z-score = -1.84, R) >droSec1.super_7 336220 111 + 3727775 GUCCUCCAUUUCCCUUUGCCUACGUUUCCUCCUCUCUUCCCAUUAGACUGGCCAUCACUUUUGAGGUUUUAUCCUUGGGCUGUCAACACGUGAUUUGACACGCCCCCUUUA ..((((...........(((...((((.................)))).)))..........))))..........((((((((((........)))))).))))...... ( -19.83, z-score = -1.34, R) >droSim1.chr2L 16385408 111 + 22036055 GUCCUCCAUUUCCCUUUGCCUACGUUUCCUCCUCUCUUCCCAUUAGGCUGGCCAUCACUUUUGAGGUUUUAUCCUUGGGCUGUCAACACGUGCUUUGACACGCCCCCUUUA ............((...(((((.....................))))).))...........((((......))))((((((((((........)))))).))))...... ( -22.30, z-score = -1.56, R) >consensus GUCCUCCAUUUCCCUUUGCCUACGUUUCCUCCUCUCUUCCCAUUAGGCUGGCCAUCACUUUUGAGGUUUUAUCCUUGGGCUGUCAACACGUGCUUUGACACGCCCCCUUUA .................(((((.....................)))))..............((((......))))((((((((((........)))))).))))...... (-19.32 = -19.52 + 0.20)


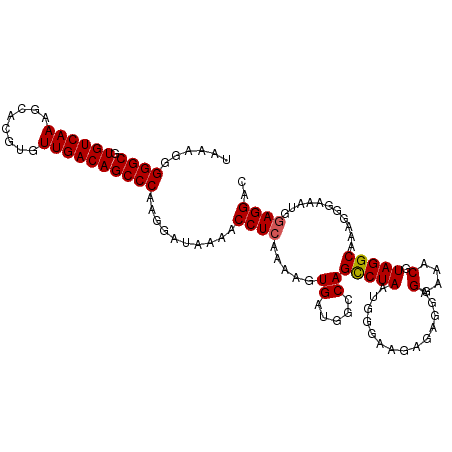
| Location | 16,689,120 – 16,689,231 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 94.95 |
| Shannon entropy | 0.08904 |
| G+C content | 0.49730 |
| Mean single sequence MFE | -27.79 |
| Consensus MFE | -25.18 |
| Energy contribution | -25.22 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.765569 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16689120 111 - 23011544 UAAAGGGGGCGUGUCAAAGCACGUGUUGACAGCCCAAGGAUAAAACCUCAAAUGUGAUGGCCAGCCUAAUGGAAAGAGUGGAGGAAACGUAGGCAAAGGGAAAUGGAGGAC ......((((.((((((........))))))))))..........((((..........(((..((....))..........(....)...)))...........)))).. ( -28.90, z-score = -1.58, R) >droEre2.scaffold_4845 19758985 111 - 22589142 UAAAGGGGGCGUGUCAAAGCACGUGUUGACAGCCCAAGGAUAAAACCUCAAAAGUGAUGGGCAGCCUAAUGGGAAGAGAGGAGGAAACGUAGGCGAUGGGAAAUGCAGGAC ......((((.((((((........))))))))))..........(((((....))).))(((.((((..............(....)(....)..))))...)))..... ( -25.70, z-score = -0.67, R) >droYak2.chr2R 16725363 111 + 21139217 UUAAAGGGGCGUGUCAAAGCACGUGUUGACAGCCCAAGGAUAAAACCUCAAAAGUGAUGGGCAGCCUAAUGGAAGGAGAGGAGGAAACGUAGGCAAAGGAAAAUGGAGGAC ......((((.((((((........))))))))))..........((((....((.....)).(((((..............(....).)))))...........)))).. ( -27.36, z-score = -1.97, R) >droSec1.super_7 336220 111 - 3727775 UAAAGGGGGCGUGUCAAAUCACGUGUUGACAGCCCAAGGAUAAAACCUCAAAAGUGAUGGCCAGUCUAAUGGGAAGAGAGGAGGAAACGUAGGCAAAGGGAAAUGGAGGAC ......((((.((((((........))))))))))..........((((..........(((..(((....)))........(....)...)))...........)))).. ( -27.90, z-score = -1.95, R) >droSim1.chr2L 16385408 111 - 22036055 UAAAGGGGGCGUGUCAAAGCACGUGUUGACAGCCCAAGGAUAAAACCUCAAAAGUGAUGGCCAGCCUAAUGGGAAGAGAGGAGGAAACGUAGGCAAAGGGAAAUGGAGGAC ......((((.((((((........))))))))))..........((((..........(((..((....))..........(....)...)))...........)))).. ( -29.10, z-score = -1.91, R) >consensus UAAAGGGGGCGUGUCAAAGCACGUGUUGACAGCCCAAGGAUAAAACCUCAAAAGUGAUGGCCAGCCUAAUGGGAAGAGAGGAGGAAACGUAGGCAAAGGGAAAUGGAGGAC ......((((.((((((........))))))))))..........((((.....((.....))(((((..............(....).)))))...........)))).. (-25.18 = -25.22 + 0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:46:01 2011