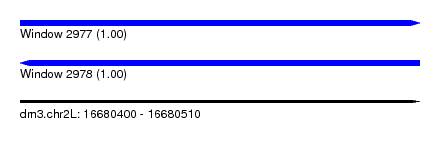
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,680,400 – 16,680,510 |
| Length | 110 |
| Max. P | 0.997547 |

| Location | 16,680,400 – 16,680,510 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 88.73 |
| Shannon entropy | 0.19899 |
| G+C content | 0.43785 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -25.72 |
| Energy contribution | -25.32 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.29 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.12 |
| SVM RNA-class probability | 0.997547 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16680400 110 + 23011544 CUUUUUGGCGCAAUAUUAUUCACCUUGACCACGCACCGGUGUGCAAAAGAUUCACCGCACACCGGUGCACUUAAAUUCACCACAGCCUUAAUAAUUCUUACGGCAUUUCC .....(((.(.(((.....((.....))....((((((((((((............))))))))))))......)))).)))..(((.(((......))).)))...... ( -31.40, z-score = -4.59, R) >droSim1.chr2L 16368501 110 + 22036055 CUUUUUGGCGCAAUAUUAUUCACCUUGACCACGCACCGGUGUGCAAAAGAUUCACCGCACAUCGGUGCAUUUAAAUUCACCACAGCCCUAAUAAUUCCUACGGCAUUUCU .....(((.(.(((.....((.....))....((((((((((((............))))))))))))......)))).)))..(((.((........)).)))...... ( -27.90, z-score = -3.25, R) >droSec1.super_7 327380 110 + 3727775 CUUUUUGGCGCAAUAUUAUUCACCUUGACCACGCACCGGUGUGCAAAAGAUUAACCGCACAUCGGUGCAUUUAAAUUCACCACAGCCGUAAUAAUUCCUACGGCAUUUCU .....(((.(.(((.....((.....))....((((((((((((............))))))))))))......)))).)))..((((((........))))))...... ( -33.60, z-score = -4.71, R) >droYak2.chr2R 16716057 108 - 21139217 CUUUUUGGCGCAAUAUUAUUCACCUUGACCACACACCGGUGUGCAAAAA-UUCACCGCACACCGGUAUAUUGAAAUUCACCACAGUCCUAAUGAUUCU-AUGAGGUUUUU ..........................((((....((((((((((.....-......))))))))))..........(((....((((.....))))..-.)))))))... ( -23.00, z-score = -1.46, R) >droEre2.scaffold_4845 19749944 110 + 22589142 CUUUUUGGCGCAAUAAUAUUCACCUUGACCAUACACCGGUGUGCAAAAAAUUCACCGCACGCCGGUGCAUACAAAUUCACCACCGCCGUAAUGAUUCUUACGCGAUUUUU ......((((.........((.....)).....(((((((((((............)))))))))))................))))((((......))))......... ( -27.60, z-score = -2.43, R) >consensus CUUUUUGGCGCAAUAUUAUUCACCUUGACCACGCACCGGUGUGCAAAAGAUUCACCGCACACCGGUGCAUUUAAAUUCACCACAGCCCUAAUAAUUCUUACGGCAUUUCU ......(((..........((.....))....((((((((((((............))))))))))))................)))....................... (-25.72 = -25.32 + -0.40)

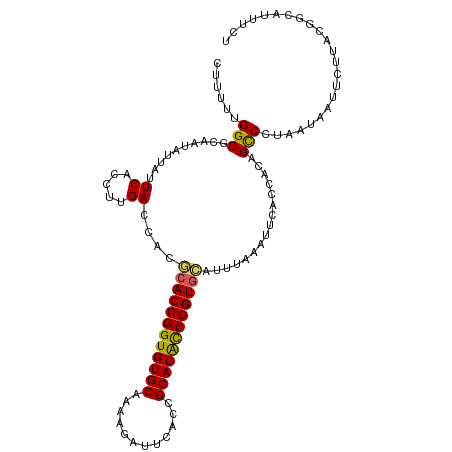
| Location | 16,680,400 – 16,680,510 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 88.73 |
| Shannon entropy | 0.19899 |
| G+C content | 0.43785 |
| Mean single sequence MFE | -37.02 |
| Consensus MFE | -32.04 |
| Energy contribution | -31.96 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.995457 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16680400 110 - 23011544 GGAAAUGCCGUAAGAAUUAUUAAGGCUGUGGUGAAUUUAAGUGCACCGGUGUGCGGUGAAUCUUUUGCACACCGGUGCGUGGUCAAGGUGAAUAAUAUUGCGCCAAAAAG .....((.(((((..((((((...(((....(((......((((((((((((((((........))))))))))))))))..))).))).)))))).))))).))..... ( -38.20, z-score = -3.27, R) >droSim1.chr2L 16368501 110 - 22036055 AGAAAUGCCGUAGGAAUUAUUAGGGCUGUGGUGAAUUUAAAUGCACCGAUGUGCGGUGAAUCUUUUGCACACCGGUGCGUGGUCAAGGUGAAUAAUAUUGCGCCAAAAAG ......(((.(((......))).)))..(((((.......((((((((.(((((((........))))))).))))))))..((.....)).........)))))..... ( -34.70, z-score = -2.26, R) >droSec1.super_7 327380 110 - 3727775 AGAAAUGCCGUAGGAAUUAUUACGGCUGUGGUGAAUUUAAAUGCACCGAUGUGCGGUUAAUCUUUUGCACACCGGUGCGUGGUCAAGGUGAAUAAUAUUGCGCCAAAAAG ......(((((((......)))))))..(((((.......((((((((.(((((((........))))))).))))))))..((.....)).........)))))..... ( -38.30, z-score = -3.33, R) >droYak2.chr2R 16716057 108 + 21139217 AAAAACCUCAU-AGAAUCAUUAGGACUGUGGUGAAUUUCAAUAUACCGGUGUGCGGUGAA-UUUUUGCACACCGGUGUGUGGUCAAGGUGAAUAAUAUUGCGCCAAAAAG ....(((....-.(((((((((......))))))..))).((((((((((((((((....-...)))))))))))))))))))...((((..........))))...... ( -35.10, z-score = -3.42, R) >droEre2.scaffold_4845 19749944 110 - 22589142 AAAAAUCGCGUAAGAAUCAUUACGGCGGUGGUGAAUUUGUAUGCACCGGCGUGCGGUGAAUUUUUUGCACACCGGUGUAUGGUCAAGGUGAAUAUUAUUGCGCCAAAAAG .......((((((...(((((((....))))))).((((((((((((((.((((((........)))))).))))))))))..))))..........))))))....... ( -38.80, z-score = -3.02, R) >consensus AGAAAUGCCGUAAGAAUUAUUACGGCUGUGGUGAAUUUAAAUGCACCGGUGUGCGGUGAAUCUUUUGCACACCGGUGCGUGGUCAAGGUGAAUAAUAUUGCGCCAAAAAG .......................(((.(..(((.......((((((((((((((((........))))))))))))))))..((.....))....)))..))))...... (-32.04 = -31.96 + -0.08)

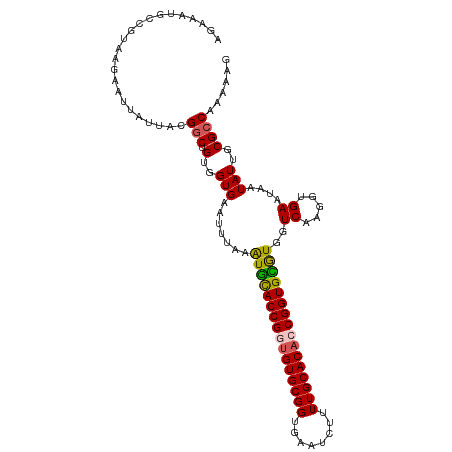
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:58 2011