
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,650,344 – 16,650,453 |
| Length | 109 |
| Max. P | 0.740968 |

| Location | 16,650,344 – 16,650,453 |
|---|---|
| Length | 109 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.36 |
| Shannon entropy | 0.55086 |
| G+C content | 0.48293 |
| Mean single sequence MFE | -18.24 |
| Consensus MFE | -12.20 |
| Energy contribution | -12.78 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.13 |
| Mean z-score | -0.66 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.740968 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16650344 109 + 23011544 ------UACCGCCGCG-UGGUCACACACAACCAC-ACGAUAACCUCCCCACAAUCCAUUUGC-CAACAUUACGCAUACGCCGCGUGGUGUCCAUAAAAAAUGCAACUCUGCAUUGUUU ------......((.(-((((........)))))-.))...................((((.-..(((((((((.......)))))))))...)))).((((((....)))))).... ( -24.40, z-score = -1.54, R) >droSim1.chr2L 16348859 111 + 22036055 -----ACACUGCCGCAUUCGUCACACUCAACCAC-ACGAUAACCUCCCCACAAUCCAUUUGC-CAACAUUACGCAUACGCCGCGUGGUGUCCAUAAAAAAUGCAACUCUGCAUUGUUU -----(((.(((.....((((.............-))))................(((((..-..(((((((((.......))))))))).......))))).......))).))).. ( -16.62, z-score = -0.22, R) >droSec1.super_7 297344 110 + 3727775 ------UACCGCUGUACUCGUCACACUCAACCAC-ACGAUAACCUCCCCACAAUCCAUUUGC-CAACAUUACGCAUACGCCGCGUGGUGUCCAUAAAAAAUGCAACUCUGCAUUGUUU ------...........((((.............-))))..................((((.-..(((((((((.......)))))))))...)))).((((((....)))))).... ( -16.32, z-score = -0.37, R) >droYak2.chr2R 16685768 102 - 21139217 --------------CUCCCUCGCAAAACAACCAC-ACGAUAACCUGUCCACAAUCCAUUUGC-CAACAUUACGCAUACGCCGCGUGGUGUCCAUAAAAAAUGCAGCUCUGCAUAGUUU --------------.......(((((........-..(((.....))).........)))))-..(((((((((.......))))))))).........(((((....)))))..... ( -18.21, z-score = -1.15, R) >droEre2.scaffold_4845 19720957 91 + 22589142 -------------------------CACAAGCAC-ACGAUAACCUACCCGCAAUCCAUUUGC-CAACAUUACGCAUACGCCGCGUGGCGUCCAUAAAAAAUGCAACUCUGCAUUGCUU -------------------------...((((..-(((...........((((.....))))-.....((((((.......)))))))))........((((((....)))))))))) ( -17.90, z-score = -0.99, R) >dp4.chr4_group3 8391375 84 + 11692001 --------------------------------ACGAAGGCGCCUCCCC-ACGAUCCAUUAGC-CGAUAUUACGCAUACGCCGCGUGGUGUCCAUAAAAAAUGCAACGCUGCAUUGUUU --------------------------------.....(((........-...........))-)((((((((((.......)))))))))).......((((((....)))))).... ( -21.01, z-score = -0.58, R) >droPer1.super_1 9840600 85 + 10282868 --------------------------------ACGAAGGCGCCCCCCCCACGAUCCAUUAGC-CGAUAUUACGCAUACGCCGCGUGGUGUCCAUAAAAAAUGCAACGCUGCAUUGUUU --------------------------------.....(((....................))-)((((((((((.......)))))))))).......((((((....)))))).... ( -20.95, z-score = -0.78, R) >droWil1.scaffold_181132 106086 116 - 1035393 UACAAAUAUACGCGUACCCAUUCCAUUCCCUCCCAUAUGCAAUCCCAUC-CAAUCCAUUAGAACAACAUUACGCAUACGCCGCAUG-UGCCCAUAAAAAAUGCAACUCUGUUUAGUGU ...........(((((...................))))).........-.....(((((((.((.......((....)).((((.-............)))).....))))))))). ( -10.53, z-score = 0.33, R) >consensus __________________C_U__CACACAACCAC_ACGAUAACCUCCCCACAAUCCAUUUGC_CAACAUUACGCAUACGCCGCGUGGUGUCCAUAAAAAAUGCAACUCUGCAUUGUUU .................................................................(((((((((.......)))))))))........((((((....)))))).... (-12.20 = -12.78 + 0.58)

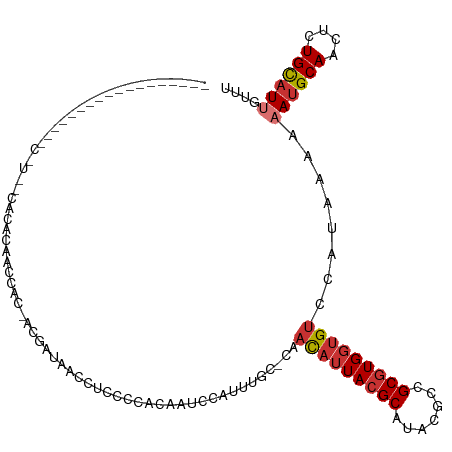
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:56 2011