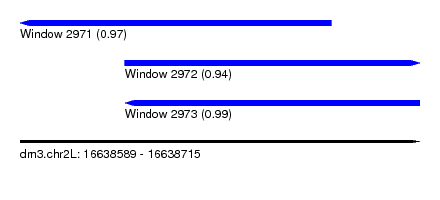
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,638,589 – 16,638,715 |
| Length | 126 |
| Max. P | 0.989093 |

| Location | 16,638,589 – 16,638,687 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 77.47 |
| Shannon entropy | 0.40789 |
| G+C content | 0.56228 |
| Mean single sequence MFE | -20.88 |
| Consensus MFE | -16.19 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973416 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
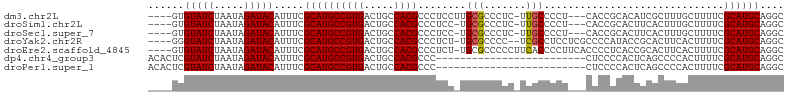
>dm3.chr2L 16638589 98 - 23011544 ----GUGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCCUUGCGCCCUC-UUGCCCCU---CACCGCACAUCGCUUUGCUUUUCGCAUGCAGGC ----(((((((.....)))))))...((((((((((.....))))........(((.....-.(((....---....)))...)))..........)))))).... ( -23.30, z-score = -1.34, R) >droSim1.chr2L 16337092 97 - 22036055 ----GUGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCC-UGCGCCCUC-UUGCCCCU---CACCGCACUUCACUUUGCUUUUCGCAUGCAGGC ----(((((((.....)))))))...((((((.((((..((..(((.....-.))).....-..))...)---))).(((........))).....)))))).... ( -22.90, z-score = -1.92, R) >droSec1.super_7 285639 97 - 3727775 ----GUGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCC-UGCGCCCUC-UUGCCCCU---CACCGCACUUCACUUUGCUUUUCGCAUGCAGGC ----(((((((.....)))))))...((((((.((((..((..(((.....-.))).....-..))...)---))).(((........))).....)))))).... ( -22.90, z-score = -1.92, R) >droYak2.chr2R 16674037 99 + 21139217 ----GGGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCU-UGCGCCCC--UCGCCUCCUCGCCCCAUACCGCACUUCACUUUUCGCAUGCAGGC ----..(((((.....))))).....((((((((((.....))))......-((((....--..((......)).......))))...........)))))).... ( -22.42, z-score = -0.68, R) >droEre2.scaffold_4845 19709006 101 - 22589142 ----GUGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCU-UGCGCCCCCUUCACCCCUUCACCCCUCACCGCACUUCACUUUUCGCAUGCAGGC ----(((((((.....)))))))...((((((((((.....))))......-((((.........................))))...........)))))).... ( -21.41, z-score = -2.86, R) >dp4.chr4_group3 8378197 81 - 11692001 ACACUCGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCC-------------------------CUCCCCACUCAGCCCCACUUUUCGCAUGCAGGC ......(((((.....))))).....((((((.(((.....)))((..-------------------------...........))..........)))))).... ( -16.62, z-score = -1.97, R) >droPer1.super_1 9827958 81 - 10282868 ACACUCGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCC-------------------------CUCCCCACUCAGCCCCACUUUUCGCAUGCAGGC ......(((((.....))))).....((((((.(((.....)))((..-------------------------...........))..........)))))).... ( -16.62, z-score = -1.97, R) >consensus ____GUGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCCUCC_UGCGCCCUC_UUGCCCCU___CACCGCACUUCACUUCACUUUUCGCAUGCAGGC ......(((((.....))))).....((((((((((.....))))........(((.......)))..............................)))))).... (-16.19 = -16.43 + 0.24)
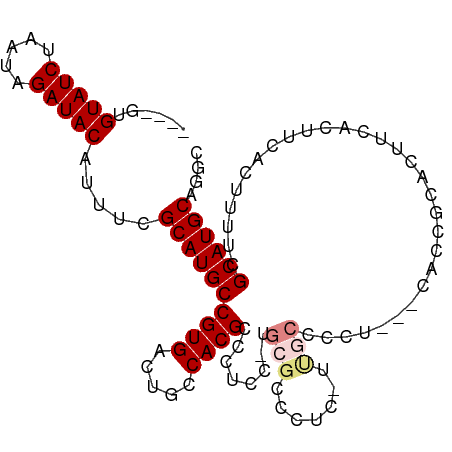

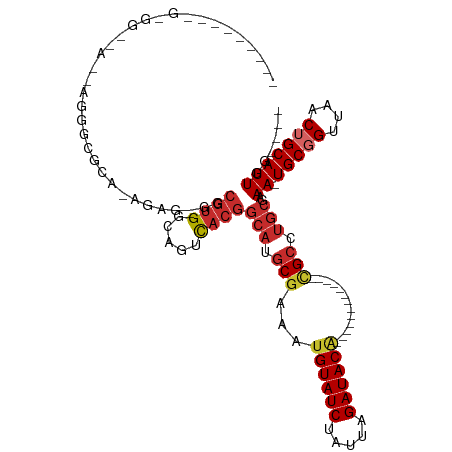
| Location | 16,638,622 – 16,638,715 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 72.00 |
| Shannon entropy | 0.48786 |
| G+C content | 0.53356 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -16.12 |
| Energy contribution | -16.53 |
| Covariance contribution | 0.41 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.938384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
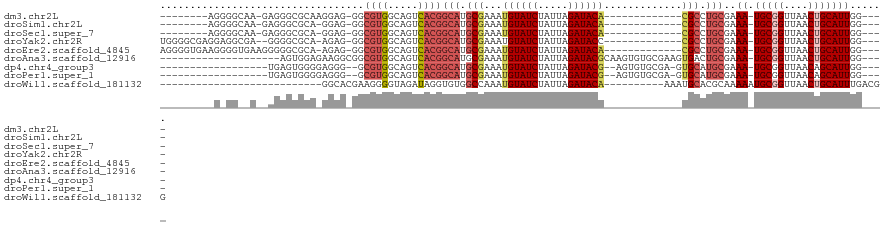
>dm3.chr2L 16638622 93 + 23011544 --------AGGGGCAA-GAGGGCGCAAGGAG-GGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACA-------------CGCCUGCGAAA-UGCGGUUAACUGCAUUGG---- --------..((((..-.....((((.....-(.((((.....)))).).))))...((((((.....))))))-------------.))))....((-(((((....)))))))..---- ( -30.10, z-score = -1.44, R) >droSim1.chr2L 16337125 92 + 22036055 --------AGGGGCAA-GAGGGCGCA-GGAG-GGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACA-------------CGCCUGCGAAA-UGCGGUUAACUGCAUUGG---- --------........-.....((((-((.(-.(((((.(......).)))))....((((((.....))))))-------------).)))))).((-(((((....)))))))..---- ( -33.40, z-score = -2.64, R) >droSec1.super_7 285672 92 + 3727775 --------AGGGGCAA-GAGGGCGCA-GGAG-GGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACA-------------CGCCUGCGAAA-UGCGGUUAACUGCAUUGG---- --------........-.....((((-((.(-.(((((.(......).)))))....((((((.....))))))-------------).)))))).((-(((((....)))))))..---- ( -33.40, z-score = -2.64, R) >droYak2.chr2R 16674065 99 - 21139217 UGGGGCGAGGAGGCGA--GGGGCGCA-AGAG-GGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACC-------------CGCCUGCGAAA-UGCGGUUAACUGCAUUGG---- ........(.(((((.--((..((((-....-(.((((.....)))).).)))).....((((.....))))))-------------))))).)..((-(((((....)))))))..---- ( -33.70, z-score = -0.70, R) >droEre2.scaffold_4845 19709034 101 + 22589142 AGGGGUGAAGGGGUGAAGGGGGCGCA-AGAG-GGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACA-------------CGCCUGCGAAA-UGCGGUUAACUGCAUUGG---- ............((((..(..((((.-....-.))))..)..)))).(((.(((...((((((.....))))))-------------))).)))..((-(((((....)))))))..---- ( -33.00, z-score = -2.13, R) >droAna3.scaffold_12916 257583 96 - 16180835 --------------------AGUGGAGAAGGCGGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACGCAAGUGUGCGAAGUGACUGCGAAA-UGCGGUUAACUGCAUUGG---- --------------------((((.((..(((.((((.((((((((.((((((....((((((.....))))))...))))))...))))))))...)-))).)))..)).))))..---- ( -39.80, z-score = -4.47, R) >dp4.chr4_group3 8378219 93 + 11692001 ------------------UGAGUGGGGAGGG--GCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACG--AGUGUGCGA-GUGCAUGCGAAA-UGCGGUUAACAGCAUUGG---- ------------------.............--(((((.((.((...((((((....((((((.....))))))--.))))))))-.)))))))..((-(((........)))))..---- ( -26.60, z-score = -0.85, R) >droPer1.super_1 9827980 93 + 10282868 ------------------UGAGUGGGGAGGG--GCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACG--AGUGUGCGA-GUGCAUGCGAAA-UGCGGUUAACAGCAUUGG---- ------------------.............--(((((.((.((...((((((....((((((.....))))))--.))))))))-.)))))))..((-(((........)))))..---- ( -26.60, z-score = -0.85, R) >droWil1.scaffold_181132 92290 84 - 1035393 ---------------------------GGCACGAAGGGGUAGAUAGGUGUGGCCAAAUGUAUCUAUUAGAUACA----------AAAUGCACGCAAAAAUGCGGUUAACUGCAUUUGACGG ---------------------------.((((....)((((((((.((........)).)))))))).......----------...))).((...((((((((....))))))))..)). ( -22.00, z-score = -1.96, R) >consensus _________G_GG__A__AGGGCGCA_AGAG_GGCGUGGCAGUCACGGCAUGCGAAAUGUAUCUAUUAGAUACA_____________CGCCUGCGAAA_UGCGGUUAACUGCAUUGG____ .....................((..........(((((.(......).))))).....(((((.....)))))...................)).....(((((....)))))........ (-16.12 = -16.53 + 0.41)

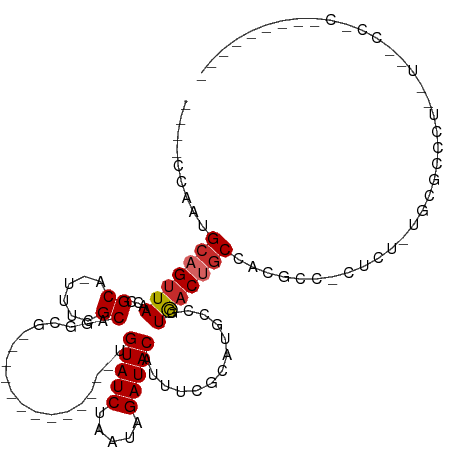
| Location | 16,638,622 – 16,638,715 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 72.00 |
| Shannon entropy | 0.48786 |
| G+C content | 0.53356 |
| Mean single sequence MFE | -22.44 |
| Consensus MFE | -12.98 |
| Energy contribution | -13.32 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.35 |
| SVM RNA-class probability | 0.989093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16638622 93 - 23011544 ----CCAAUGCAGUUAACCGCA-UUUCGCAGGCG-------------UGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCC-CUCCUUGCGCCCUC-UUGCCCCU-------- ----.....(((((((...(((-(...((.((.(-------------((((((.....))))))).))))))))..)))))))..(((.-......))).....-........-------- ( -23.90, z-score = -1.99, R) >droSim1.chr2L 16337125 92 - 22036055 ----CCAAUGCAGUUAACCGCA-UUUCGCAGGCG-------------UGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCC-CUCC-UGCGCCCUC-UUGCCCCU-------- ----.....(((((((...(((-(...((.((.(-------------((((((.....))))))).))))))))..)))))))..(((.-....-.))).....-........-------- ( -24.40, z-score = -2.17, R) >droSec1.super_7 285672 92 - 3727775 ----CCAAUGCAGUUAACCGCA-UUUCGCAGGCG-------------UGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCC-CUCC-UGCGCCCUC-UUGCCCCU-------- ----.....(((((((...(((-(...((.((.(-------------((((((.....))))))).))))))))..)))))))..(((.-....-.))).....-........-------- ( -24.40, z-score = -2.17, R) >droYak2.chr2R 16674065 99 + 21139217 ----CCAAUGCAGUUAACCGCA-UUUCGCAGGCG-------------GGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCC-CUCU-UGCGCCCC--UCGCCUCCUCGCCCCA ----..(((((........)))-))..(.(((((-------------((((((.....)))))....((((.(.((((.....)))).)-....-))))....--)))))).)........ ( -24.30, z-score = -0.48, R) >droEre2.scaffold_4845 19709034 101 - 22589142 ----CCAAUGCAGUUAACCGCA-UUUCGCAGGCG-------------UGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCC-CUCU-UGCGCCCCCUUCACCCCUUCACCCCU ----.....(((((((...(((-(...((.((.(-------------((((((.....))))))).))))))))..)))))))..(((.-....-.)))...................... ( -24.40, z-score = -3.13, R) >droAna3.scaffold_12916 257583 96 + 16180835 ----CCAAUGCAGUUAACCGCA-UUUCGCAGUCACUUCGCACACUUGCGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCCGCCUUCUCCACU-------------------- ----..(((((........)))-))..((((((((...(((....)))(((((.....)))))............))))))))..................-------------------- ( -25.00, z-score = -3.25, R) >dp4.chr4_group3 8378219 93 - 11692001 ----CCAAUGCUGUUAACCGCA-UUUCGCAUGCAC-UCGCACACU--CGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGC--CCCUCCCCACUCA------------------ ----..(((((.(....).)))-))..((((((..-..(......--)(((((.....))))).....))))))((((.....)))).--.............------------------ ( -19.40, z-score = -2.49, R) >droPer1.super_1 9827980 93 - 10282868 ----CCAAUGCUGUUAACCGCA-UUUCGCAUGCAC-UCGCACACU--CGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGC--CCCUCCCCACUCA------------------ ----..(((((.(....).)))-))..((((((..-..(......--)(((((.....))))).....))))))((((.....)))).--.............------------------ ( -19.40, z-score = -2.49, R) >droWil1.scaffold_181132 92290 84 + 1035393 CCGUCAAAUGCAGUUAACCGCAUUUUUGCGUGCAUUU----------UGUAUCUAAUAGAUACAUUUGGCCACACCUAUCUACCCCUUCGUGCC--------------------------- ..((((((((((......((((....))))))))...----------((((((.....))))))))))))........................--------------------------- ( -16.80, z-score = -1.98, R) >consensus ____CCAAUGCAGUUAACCGCA_UUUCGCAGGCG_____________UGUAUCUAAUAGAUACAUUUCGCAUGCCGUGACUGCCACGCC_CUCU_UGCGCCCU__U__CC_C_________ .........(((((((...((......))...................(((((.....))))).............)))))))...................................... (-12.98 = -13.32 + 0.35)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:54 2011