
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,636,582 – 16,636,706 |
| Length | 124 |
| Max. P | 0.689136 |

| Location | 16,636,582 – 16,636,674 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 66.60 |
| Shannon entropy | 0.62006 |
| G+C content | 0.49100 |
| Mean single sequence MFE | -25.30 |
| Consensus MFE | -8.40 |
| Energy contribution | -9.10 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.575819 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
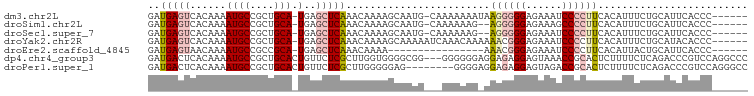
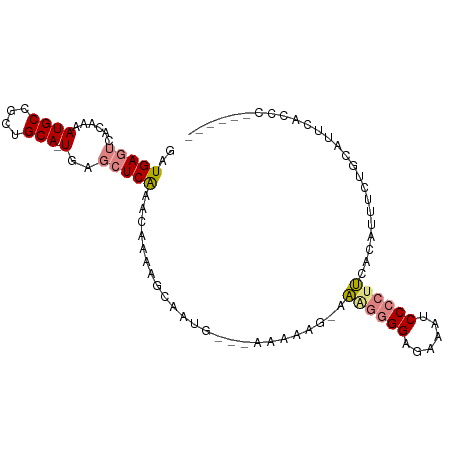
>dm3.chr2L 16636582 92 + 23011544 GAUGAGUCACAAAAUGCCGCUGCA-UGAGCUCAAACAAAAGCAAUG-CAAAAAAAUAAGGGGGAGAAAUCCCCUUCACAUUUCUGCAUUCACCC------ ..(((((......((((....)))-)..))))).......(.((((-((.(((....(((((((....)))))))....))).)))))))....------ ( -25.90, z-score = -3.05, R) >droSim1.chr2L 16335141 90 + 22036055 GAUGAGUCACAAAAUGCCGCUGCA-UGAGCUCAAACAAAAGCAAUG-CAAAAAAG--AGGGGGAGAAAGCCCCUUCACAUUUCUGCAUUCACCC------ ..(((((((..(((((....((((-(..(((........))).)))-)).....(--((((((......))))))).))))).)).)))))...------ ( -25.30, z-score = -2.06, R) >droSec1.super_7 283686 90 + 3727775 GAUGAGUCACAAAAUGCCGCUGCA-UGAGCUCAAACAAAAGCAAUG-CAAAAAAG--AGGGGGAGAAAUCCCCUUCACAUUUCUGCAUUCACCC------ ..(((((((..(((((....((((-(..(((........))).)))-)).....(--(((((((....)))))))).))))).)).)))))...------ ( -29.30, z-score = -3.77, R) >droYak2.chr2R 16672214 93 - 21139217 GAUGAGUCACAAAAUGCCGCUGCA-UGAGCUCAAACAAAAGCAAAAAUCAAACAAAAAACGGGAGAAAUCCCCUUCACAUUUCUGCAUACACCC------ ..(((((......((((....)))-)..))))).......(((.((((............((((....))))......)))).)))........------ ( -15.17, z-score = -1.26, R) >droEre2.scaffold_4845 19707216 77 + 22589142 GAUGAGUAACAAAAUGCCGCCGCA-UGAGCUCAAACAAAA----------------AAACGGGAGAAAUCCCCUUCACAUUACUGCAUUCACCC------ (((((((((....((((....)))-)..............----------------....((((....)))).......))))).)))).....------ ( -14.90, z-score = -2.01, R) >dp4.chr4_group3 8375614 97 + 11692001 GAUGACUCACAAAAUGCCGCUGCACUGUUCUCGCUUGGUGGGGCGG---GGGGGGAGGAGAGGAGUAAACCGCACUCUUUUCUCAGACCCGUCCAGGCCC ...............(((....(((((........)))))((((((---(..((((((((((..((.....)).))))))))))...))))))).))).. ( -37.20, z-score = -0.48, R) >droPer1.super_1 9825381 92 + 10282868 GAUGACUCACAAAAUGCCGCUGCACUGUUCUCGCUUGGGGGAG--------GGGGAGGAGAGGAGUAGACCGCACUCUUUUCUCAGACCCGUCCAGGGCC ...............(((.(((.((....(((.(....).)))--------(((...((((((((.((......))))))))))...))))).)))))). ( -29.30, z-score = 0.93, R) >consensus GAUGAGUCACAAAAUGCCGCUGCA_UGAGCUCAAACAAAAGCAAUG___AAAAAG_AAAGGGGAGAAAUCCCCUUCACAUUUCUGCAUUCACCC______ ..(((((..((...(((....))).)).)))))........................((((((......))))))......................... ( -8.40 = -9.10 + 0.70)
| Location | 16,636,610 – 16,636,706 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 59.92 |
| Shannon entropy | 0.65815 |
| G+C content | 0.52179 |
| Mean single sequence MFE | -34.30 |
| Consensus MFE | -14.36 |
| Energy contribution | -13.52 |
| Covariance contribution | -0.84 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.689136 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
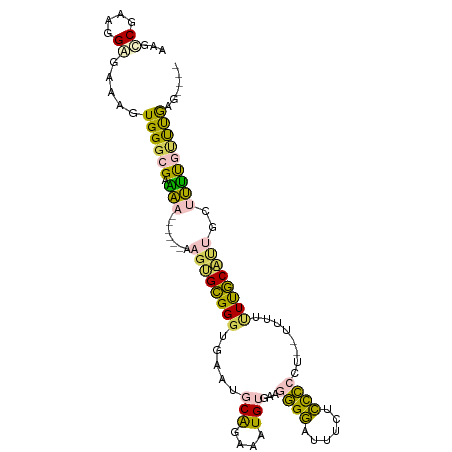
>dm3.chr2L 16636610 96 - 23011544 AAGACGAAGGAGAAAGUGGGCGGAAA------AAGUGCGGGUGAAUGCAGAAAUGUGAAGGGGAUUUCUCCCCCUUAUUUUUUUGCAUUGCUUUUGUUUGAG---- .(((((((((......((.((.....------..)).))....((((((((((.((((.(((((....))))).)))).)))))))))).)))))))))...---- ( -31.10, z-score = -2.71, R) >droSim1.chr2L 16335169 94 - 22036055 AAGUCGCAGGUGAAAGUGGGCGUAAA------AAGUGCGGGUGAAUGCAGAAAUGUGAAGGGGCUUUCUCCCCCU--CUUUUUUGCAUUGCUUUUGUUUGAG---- ......((((..(((((...((((..------...))))....((((((((((...((.((((......)))).)--).)))))))))))))))..))))..---- ( -29.50, z-score = -1.51, R) >droSec1.super_7 283714 94 - 3727775 AAGUCGCAGGAGAAAGUGGGCGUAAA------AAGUGCGGGUGAAUGCAGAAAUGUGAAGGGGAUUUCUCCCCCU--CUUUUUUGCAUUGCUUUUGUUUGAG---- ......((((.((((((...((((..------...))))....((((((((((...((.(((((....))))).)--).)))))))))))))))).))))..---- ( -28.60, z-score = -1.48, R) >dp4.chr4_group3 8375643 106 - 11692001 AAACCGAAAGGGAAAGUGGGCGAGGGAGAGCUAAGGGGGGGCCUGGACGGGUCUGAGAAAAGAGUGCGGUUUACUCCUCUCCUCCCCCCCCGCCCCACCAAGCGAG ...((....))....(((((((.(((........((((((((((....))))))((((...(((((.....))))).))))..))))))))).)))))........ ( -41.00, z-score = 0.66, R) >droPer1.super_1 9825410 101 - 10282868 AAACCGAAAGGGAAAGUGGGCGAGGGAGAGCUAAGAGGGGCCCUGGACGGGUCUGAGAAAAGAGUGCGGUCUACUCCUCUCCUCCCC-----CUCCCCCAAGCGAG ...((....)).....((((.(((((.(((...(((((((...(((((.(.(((......)))...).)))))))))))).))).))-----))).))))...... ( -41.30, z-score = -0.54, R) >consensus AAGCCGAAGGAGAAAGUGGGCGAAAA______AAGUGCGGGUGAAUGCAGAAAUGUGAAGGGGAUUUCUCCCCCU__UUUUUUUGCAUUGCUUUUGUUUGAG____ ...((....)).......................(.(((((..((((((((((.(.((.((((......)))).)).).))))))))))...))))).)....... (-14.36 = -13.52 + -0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:51 2011