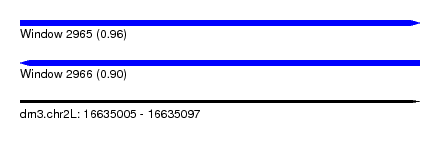
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,635,005 – 16,635,097 |
| Length | 92 |
| Max. P | 0.955916 |

| Location | 16,635,005 – 16,635,097 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 64.41 |
| Shannon entropy | 0.73881 |
| G+C content | 0.57119 |
| Mean single sequence MFE | -20.77 |
| Consensus MFE | -9.42 |
| Energy contribution | -9.96 |
| Covariance contribution | 0.54 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955916 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
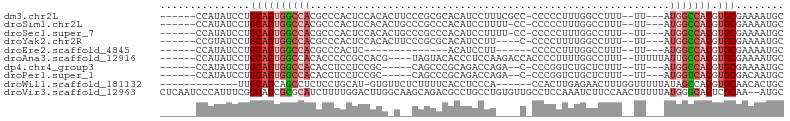
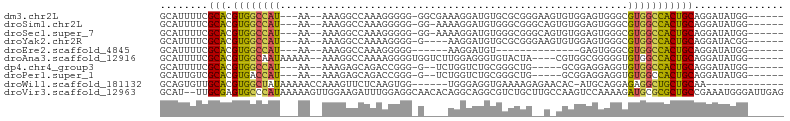
>dm3.chr2L 16635005 92 + 23011544 ------CCAUAUCCUGCAGUGGCCACGCCCACUCCACACUUCCCGCGCACAUCCUUUCGCC-CCCCCUUUGGCCUUU--UU---AUGGCCACGUGCGAAAAUGC ------........((((((((((((((................)))...........(((-........)))....--..---.))))))).))))....... ( -21.29, z-score = -1.56, R) >droSim1.chr2L 16333582 91 + 22036055 ------CCAUAUCCUGCAGUGGCCACGCCCACUCCACACUGCCCGCCCACAUCCUUUU-CC-CCCCCUUUGGCCUUU--UU---AUGGCCACGUGCGAAAAUGC ------........(((((((((((.((............))..(((...........-..-........)))....--..---.))))))).))))....... ( -17.60, z-score = -0.66, R) >droSec1.super_7 282139 91 + 3727775 ------CCAUAUCCUGCAGUGGCCACGCCCACUCCACACUGCCCGCCCACAUCCUUUU-CC-CCCCCUUUGGCCUUU--UU---AUGGCCACGUGCGAAAAUGC ------........(((((((((((.((............))..(((...........-..-........)))....--..---.))))))).))))....... ( -17.60, z-score = -0.66, R) >droYak2.chr2R 16670631 88 - 21139217 ------CCGUAUCCUGCAGUGGCCACGCCCACUCCACACUUCCCGCGCACAUCCUU----C-CCCCUUUUGGCCUUU--UU---AUGGCCACGUGCGAAAAUGC ------..((((..((((((((((((((................))).........----.-..((....)).....--..---.))))))).))))...)))) ( -19.49, z-score = -1.01, R) >droEre2.scaffold_4845 19705683 73 + 22589142 ------CCAUAUCCUGCAGUGGCCACGCCCACUC--------------ACAUCCUU------CCCCCUUUGGCCUUU--UU---AUGGCCACGUGCGAAAAUGC ------........(((((((((((.(((.....--------------........------........)))....--..---.))))))).))))....... ( -16.20, z-score = -0.94, R) >droAna3.scaffold_12916 254619 92 - 16180835 ------CCAUAUCCUGCAGUGGCCACACCCCCGCCACG----UAGUACACCCUCCAAGACCACCCCUUUUGGCCUUU--UUUUUAUUGCCACGUGCGAAAAUGC ------.........((((((....)))...(((.(((----(.(((........(((.(((.......))).))).--.......))).)))))))....))) ( -16.19, z-score = -0.76, R) >dp4.chr4_group3 8374265 85 + 11692001 ------CCAUAUCCUGCAGUGGCCACACCUCCUCCGC-----CAGCCCGCAGACCAGA--C-CCCGGUCUGCUCUUU--UU---AUGGCCACGUGCGAAAAUGC ------........(((((((((((..........(.-----...)..(((((((...--.-...))))))).....--..---.))))))).))))....... ( -28.20, z-score = -3.07, R) >droPer1.super_1 9824053 85 + 10282868 ------CCAUAUCCUGCAGUGGCCACACCUCCUCCGC-----CAGCCCGCAGACCAGA--C-CCCGGUCUGCUCUUU--UU---AUGGUCACGUGCGACAAUGC ------........(((((((((((..........(.-----...)..(((((((...--.-...))))))).....--..---.))))))).))))....... ( -25.50, z-score = -1.92, R) >droWil1.scaffold_181132 88215 84 - 1035393 -------------UUGCAGCAGCCUCUCCUGCAU-GUGUUCUCUUUUCACCUCCCA------CCACUUGAGAACUUUGGUUUUUAUAGCCACGUGCAACACUGC -------------.....((((.......(((((-((((((((.............------......))))))...((((.....)))))))))))...)))) ( -20.41, z-score = -2.15, R) >droVir3.scaffold_12963 7305566 102 - 20206255 CUCAAUCCCAUUUCGGCAGCGCGCAUCUUUUGGACUUGGCAAGCAGACGCCUGCCUGUGUUGCCUCCAAAUCUUCCAACUUUUUAUGGGCACUCGCAA--AUGC ........((((((((((((((.((.....)).....((((.((....)).)))).)))))))).........((((........)))).....).))--))). ( -25.20, z-score = 0.19, R) >consensus ______CCAUAUCCUGCAGUGGCCACGCCCACUCCACA_U__CCGCCCACAUCCCU____C_CCCCCUUUGGCCUUU__UU___AUGGCCACGUGCGAAAAUGC ...............((((((((((............................................................))))))).)))........ ( -9.42 = -9.96 + 0.54)
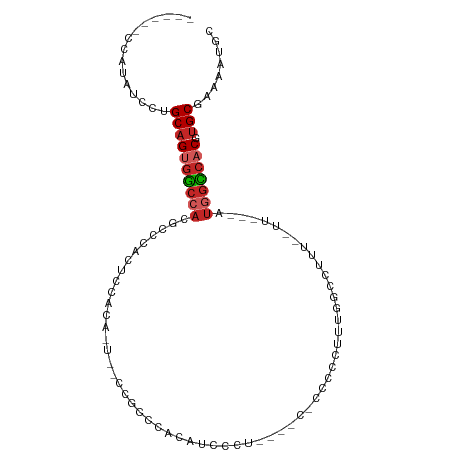
| Location | 16,635,005 – 16,635,097 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 64.41 |
| Shannon entropy | 0.73881 |
| G+C content | 0.57119 |
| Mean single sequence MFE | -28.75 |
| Consensus MFE | -12.67 |
| Energy contribution | -12.40 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.55 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.16 |
| SVM RNA-class probability | 0.901723 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
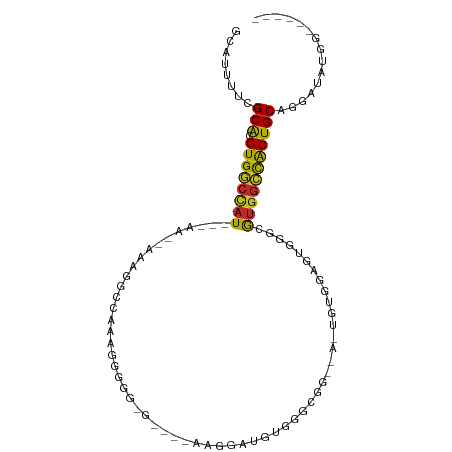
>dm3.chr2L 16635005 92 - 23011544 GCAUUUUCGCACGUGGCCAU---AA--AAAGGCCAAAGGGGG-GGCGAAAGGAUGUGCGCGGGAAGUGUGGAGUGGGCGUGGCCACUGCAGGAUAUGG------ ((((((((((((((((((..---..--...))))........-..(....).)))))))...)))))))(.(((((......))))).).........------ ( -31.00, z-score = -1.39, R) >droSim1.chr2L 16333582 91 - 22036055 GCAUUUUCGCACGUGGCCAU---AA--AAAGGCCAAAGGGGG-GG-AAAAGGAUGUGGGCGGGCAGUGUGGAGUGGGCGUGGCCACUGCAGGAUAUGG------ (((((..(.(...(((((..---..--...)))))...).).-.)-).......((((.((.((............)).)).))))))).........------ ( -28.50, z-score = -1.10, R) >droSec1.super_7 282139 91 - 3727775 GCAUUUUCGCACGUGGCCAU---AA--AAAGGCCAAAGGGGG-GG-AAAAGGAUGUGGGCGGGCAGUGUGGAGUGGGCGUGGCCACUGCAGGAUAUGG------ (((((..(.(...(((((..---..--...)))))...).).-.)-).......((((.((.((............)).)).))))))).........------ ( -28.50, z-score = -1.10, R) >droYak2.chr2R 16670631 88 + 21139217 GCAUUUUCGCACGUGGCCAU---AA--AAAGGCCAAAAGGGG-G----AAGGAUGUGCGCGGGAAGUGUGGAGUGGGCGUGGCCACUGCAGGAUACGG------ ((((((((.(.(.(((((..---..--...)))))...).).-)----)))...)))).((.....(.((.(((((......))))).)).)...)).------ ( -27.40, z-score = -0.62, R) >droEre2.scaffold_4845 19705683 73 - 22589142 GCAUUUUCGCACGUGGCCAU---AA--AAAGGCCAAAGGGGG------AAGGAUGU--------------GAGUGGGCGUGGCCACUGCAGGAUAUGG------ .(((.((((((.((((((((---..--...(.(((..(.(..------.....).)--------------...))).)))))))))))).))).))).------ ( -23.10, z-score = -1.14, R) >droAna3.scaffold_12916 254619 92 + 16180835 GCAUUUUCGCACGUGGCAAUAAAAA--AAAGGCCAAAAGGGGUGGUCUUGGAGGGUGUACUA----CGUGGCGGGGGUGUGGCCACUGCAGGAUAUGG------ (((((..(((((((((..(((....--.(((((((.......)))))))......))).)))----))).)))..)))))..((......))......------ ( -28.90, z-score = -1.51, R) >dp4.chr4_group3 8374265 85 - 11692001 GCAUUUUCGCACGUGGCCAU---AA--AAAGAGCAGACCGGG-G--UCUGGUCUGCGGGCUG-----GCGGAGGAGGUGUGGCCACUGCAGGAUAUGG------ .(((.((((((.((((((((---..--.....(((((((((.-.--.)))))))))..(((.-----.(....).)))))))))))))).))).))).------ ( -37.20, z-score = -2.85, R) >droPer1.super_1 9824053 85 - 10282868 GCAUUGUCGCACGUGACCAU---AA--AAAGAGCAGACCGGG-G--UCUGGUCUGCGGGCUG-----GCGGAGGAGGUGUGGCCACUGCAGGAUAUGG------ (((..(((((((.(..((..---..--..((.(((((((((.-.--.)))))))))...)).-----.....))).)))))))...))).........------ ( -30.00, z-score = -0.55, R) >droWil1.scaffold_181132 88215 84 + 1035393 GCAGUGUUGCACGUGGCUAUAAAAACCAAAGUUCUCAAGUGG------UGGGAGGUGAAAAGAGAACAC-AUGCAGGAGAGGCUGCUGCAA------------- (((((.(((((.(((.((......(((.....(((((.....------))))))))......))..)))-.))))....).))))).....------------- ( -20.20, z-score = 0.32, R) >droVir3.scaffold_12963 7305566 102 + 20206255 GCAU--UUGCGAGUGCCCAUAAAAAGUUGGAAGAUUUGGAGGCAACACAGGCAGGCGUCUGCUUGCCAAGUCCAAAAGAUGCGCGCUGCCGAAAUGGGAUUGAG .(((--((((.((((((((........)))....(((((((....)...(((((((....)))))))...))))))......)))))))..)))))........ ( -32.70, z-score = -0.71, R) >consensus GCAUUUUCGCACGUGGCCAU___AA__AAAGGCCAAAGGGGG_G____AAGGAUGUGGGCGG__A_UGUGGAGUGGGCGUGGCCACUGCAGGAUAUGG______ ........(((.((((((((..........................................................)))))))))))............... (-12.67 = -12.40 + -0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:48 2011