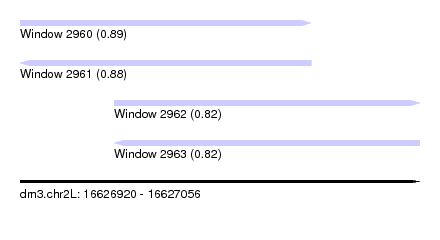
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,626,920 – 16,627,056 |
| Length | 136 |
| Max. P | 0.889465 |

| Location | 16,626,920 – 16,627,019 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 86.98 |
| Shannon entropy | 0.22602 |
| G+C content | 0.46000 |
| Mean single sequence MFE | -25.70 |
| Consensus MFE | -21.54 |
| Energy contribution | -22.14 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.889465 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
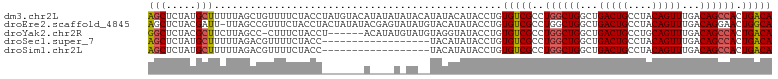
>dm3.chr2L 16626920 99 + 23011544 GGGUCGUAAAAA-GAAAAGUGCAUAAAAUGUCUUGUCAGUGGCUGUCAAACUGUAGGCAGUCAGCCAGCCAGGCGACACAGGUAUGUAUAUGUAUAUAUA ............-.....(((((((..((..((((((..((((((((........))))))))(((.....))))))).))..))...)))))))..... ( -29.10, z-score = -2.67, R) >droEre2.scaffold_4845 19697468 98 + 22589142 GGGUCGUAAAAAAGAAAAGUGCAUAAAAUGUCUUGCCAGUUCCUGUCAAACUGUAGGCAGUCAGCCAGCCAGGCGACACAGGUAUAUGUACAUAUACU-- ..................(((((((...(((((...(((((.......))))).)))))(((.(((.....)))))).......))))))).......-- ( -23.50, z-score = -0.92, R) >droYak2.chr2R 16662471 93 - 21139217 GGGUCGUAAAAA-GAAAAGUGCAUAAAAUGUCUUGUCAGUGGCUGUCAAACUGCAGGCAGUCAGCCAGCCAGGCGACACAGGUAUACCUACAUA------ ..(((((.....-((.(((.((((...))))))).))..((((((.(..((((....))))..).)))))).)))))..(((....))).....------ ( -26.90, z-score = -1.57, R) >droSec1.super_7 274096 83 + 3727775 GGGUCGUAAAAA-GAAAAGUGCAUAAAAUGUCUUGUCAGUGGCUGUCAAACUGUAGGCAGUCAGCCAGCCAGGCGACACAGGUA---------------- ..(((((.....-((.(((.((((...))))))).))..((((((.(..((((....))))..).)))))).))))).......---------------- ( -24.50, z-score = -2.00, R) >droSim1.chr2L 16322541 83 + 22036055 GGGUCGUAAAAA-GAAAAGUGCAUAAAAUGUCUUGUCAGUGGCUGUCAAACUGUAGGCAGUCAGCCAGCCAGGCGACACAGGUA---------------- ..(((((.....-((.(((.((((...))))))).))..((((((.(..((((....))))..).)))))).))))).......---------------- ( -24.50, z-score = -2.00, R) >consensus GGGUCGUAAAAA_GAAAAGUGCAUAAAAUGUCUUGUCAGUGGCUGUCAAACUGUAGGCAGUCAGCCAGCCAGGCGACACAGGUAU___UA__UA______ ..(((((......((.(((.((((...))))))).))..((((((.(..((((....))))..).)))))).)))))....................... (-21.54 = -22.14 + 0.60)


| Location | 16,626,920 – 16,627,019 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 86.98 |
| Shannon entropy | 0.22602 |
| G+C content | 0.46000 |
| Mean single sequence MFE | -21.40 |
| Consensus MFE | -18.14 |
| Energy contribution | -18.38 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.85 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.884264 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16626920 99 - 23011544 UAUAUAUACAUAUACAUACCUGUGUCGCCUGGCUGGCUGACUGCCUACAGUUUGACAGCCACUGACAAGACAUUUUAUGCACUUUUC-UUUUUACGACCC ..............((((...(((((.(.((((((...(((((....)))))...))))))..)....)))))..))))........-............ ( -21.70, z-score = -2.61, R) >droEre2.scaffold_4845 19697468 98 - 22589142 --AGUAUAUGUACAUAUACCUGUGUCGCCUGGCUGGCUGACUGCCUACAGUUUGACAGGAACUGGCAAGACAUUUUAUGCACUUUUCUUUUUUACGACCC --.((((((....))))))..((((((((.....)))....((((...(((((.....))))))))).)))))........................... ( -20.10, z-score = -0.03, R) >droYak2.chr2R 16662471 93 + 21139217 ------UAUGUAGGUAUACCUGUGUCGCCUGGCUGGCUGACUGCCUGCAGUUUGACAGCCACUGACAAGACAUUUUAUGCACUUUUC-UUUUUACGACCC ------.......(((((...(((((.(.((((((...(((((....)))))...))))))..)....)))))..))))).......-............ ( -25.20, z-score = -1.78, R) >droSec1.super_7 274096 83 - 3727775 ----------------UACCUGUGUCGCCUGGCUGGCUGACUGCCUACAGUUUGACAGCCACUGACAAGACAUUUUAUGCACUUUUC-UUUUUACGACCC ----------------...((.(((((..((((((...(((((....)))))...)))))).)))))))..................-............ ( -20.00, z-score = -2.21, R) >droSim1.chr2L 16322541 83 - 22036055 ----------------UACCUGUGUCGCCUGGCUGGCUGACUGCCUACAGUUUGACAGCCACUGACAAGACAUUUUAUGCACUUUUC-UUUUUACGACCC ----------------...((.(((((..((((((...(((((....)))))...)))))).)))))))..................-............ ( -20.00, z-score = -2.21, R) >consensus ______UA__UA___AUACCUGUGUCGCCUGGCUGGCUGACUGCCUACAGUUUGACAGCCACUGACAAGACAUUUUAUGCACUUUUC_UUUUUACGACCC ...................((.(((((..((((((...(((((....)))))...)))))).)))))))............................... (-18.14 = -18.38 + 0.24)

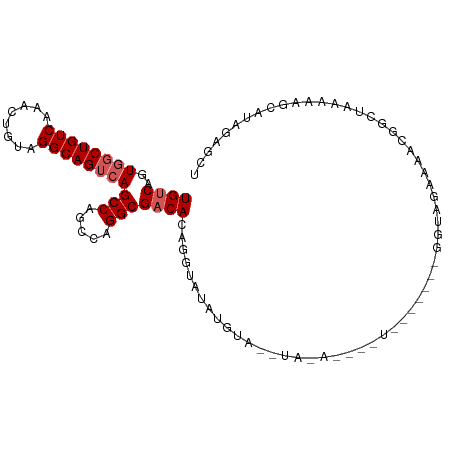
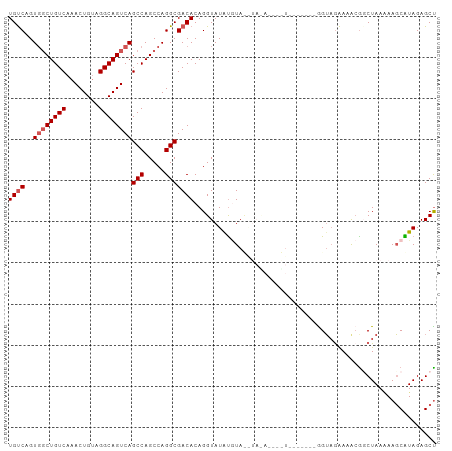
| Location | 16,626,952 – 16,627,056 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.51 |
| Shannon entropy | 0.37807 |
| G+C content | 0.46967 |
| Mean single sequence MFE | -28.96 |
| Consensus MFE | -16.56 |
| Energy contribution | -17.16 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.824711 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16626952 104 + 23011544 UGUCAGUGGCUGUCAAACUGUAGGCAGUCAGCCAGCCAGGCGACACAGGUAUGUAUAUGUAUAUAUAUGUACAUAGGUAGAAAACAGCUAAAAAGCAUAGAGCU ((((..((((((((........))))))))(((.....)))))))....(((((((((((....)))))))))))...........(((....)))........ ( -30.90, z-score = -2.09, R) >droEre2.scaffold_4845 19697501 103 + 22589142 UGCCAGUUCCUGUCAAACUGUAGGCAGUCAGCCAGCCAGGCGACACAGGUAUAUGUACAUAUACUCGUAUAUAGUAGGUAGAAACGGCUAA-AAUCGUAGAGCU .(((.(((.((..(..((((((.((.(((.(((.....))))))...(((((((....))))))).)).)))))).)..)).))))))...-............ ( -28.10, z-score = -1.27, R) >droYak2.chr2R 16662503 97 - 21139217 UGUCAGUGGCUGUCAAACUGCAGGCAGUCAGCCAGCCAGGCGACACAGGUAUACCUACAUACAUAUGU------AGGUAGAAAG-GGCUAAGAAGCGUAGAGCC ((((..((((((((........))))))))(((.....)))))))..(((.(((((((((....))))------))))).....-.(((....))).....))) ( -34.40, z-score = -2.66, R) >droSec1.super_7 274128 86 + 3727775 UGUCAGUGGCUGUCAAACUGUAGGCAGUCAGCCAGCCAGGCGACACAGGUAUAUGUA------------------GGUAGAAAACGUCUAAAAAGCAUAGAGCU ((((..((((((((........))))))))(((.....)))))))..(((.(((((.------------------..((((.....))))....)))))..))) ( -25.70, z-score = -1.98, R) >droSim1.chr2L 16322573 86 + 22036055 UGUCAGUGGCUGUCAAACUGUAGGCAGUCAGCCAGCCAGGCGACACAGGUAUAUGUA------------------GGUAGAAAACGUCUAAAAAGCAUAGAGCU ((((..((((((((........))))))))(((.....)))))))..(((.(((((.------------------..((((.....))))....)))))..))) ( -25.70, z-score = -1.98, R) >consensus UGUCAGUGGCUGUCAAACUGUAGGCAGUCAGCCAGCCAGGCGACACAGGUAUAUGUA__UA_A____U_______GGUAGAAAACGGCUAAAAAGCAUAGAGCU ((((..((((((((........))))))))(((.....)))))))........................................................... (-16.56 = -17.16 + 0.60)

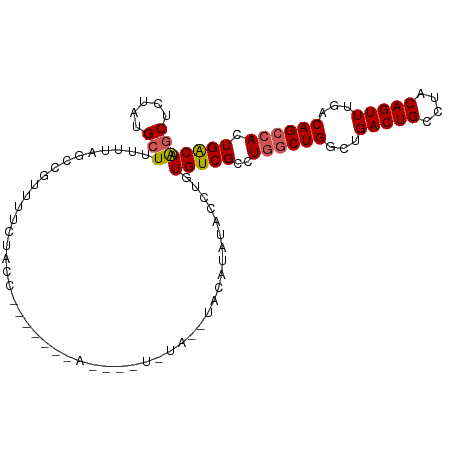
| Location | 16,626,952 – 16,627,056 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.51 |
| Shannon entropy | 0.37807 |
| G+C content | 0.46967 |
| Mean single sequence MFE | -25.96 |
| Consensus MFE | -17.36 |
| Energy contribution | -17.64 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.80 |
| SVM RNA-class probability | 0.822074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16626952 104 - 23011544 AGCUCUAUGCUUUUUAGCUGUUUUCUACCUAUGUACAUAUAUAUACAUAUACAUACCUGUGUCGCCUGGCUGGCUGACUGCCUACAGUUUGACAGCCACUGACA ........(((....)))...........((((((........))))))..........(((((..((((((...(((((....)))))...)))))).))))) ( -23.70, z-score = -0.85, R) >droEre2.scaffold_4845 19697501 103 - 22589142 AGCUCUACGAUU-UUAGCCGUUUCUACCUACUAUAUACGAGUAUAUGUACAUAUACCUGUGUCGCCUGGCUGGCUGACUGCCUACAGUUUGACAGGAACUGGCA ............-...(((((((((...........(((.((((((....)))))).)))((((.(((...(((.....)))..)))..)))))))))).))). ( -28.10, z-score = -1.24, R) >droYak2.chr2R 16662503 97 + 21139217 GGCUCUACGCUUCUUAGCC-CUUUCUACCU------ACAUAUGUAUGUAGGUAUACCUGUGUCGCCUGGCUGGCUGACUGCCUGCAGUUUGACAGCCACUGACA (((..((((((....))).-.....(((((------((((....))))))))).....)))..)))((((((...(((((....)))))...))))))...... ( -35.60, z-score = -3.37, R) >droSec1.super_7 274128 86 - 3727775 AGCUCUAUGCUUUUUAGACGUUUUCUACC------------------UACAUAUACCUGUGUCGCCUGGCUGGCUGACUGCCUACAGUUUGACAGCCACUGACA (((.....)))...((((.....))))..------------------............(((((..((((((...(((((....)))))...)))))).))))) ( -21.20, z-score = -1.38, R) >droSim1.chr2L 16322573 86 - 22036055 AGCUCUAUGCUUUUUAGACGUUUUCUACC------------------UACAUAUACCUGUGUCGCCUGGCUGGCUGACUGCCUACAGUUUGACAGCCACUGACA (((.....)))...((((.....))))..------------------............(((((..((((((...(((((....)))))...)))))).))))) ( -21.20, z-score = -1.38, R) >consensus AGCUCUAUGCUUUUUAGCCGUUUUCUACC_______A____U_UA__UACAUAUACCUGUGUCGCCUGGCUGGCUGACUGCCUACAGUUUGACAGCCACUGACA (((.....)))................................................(((((..((((((...(((((....)))))...)))))).))))) (-17.36 = -17.64 + 0.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:46 2011