
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,623,349 – 16,623,446 |
| Length | 97 |
| Max. P | 0.723789 |

| Location | 16,623,349 – 16,623,446 |
|---|---|
| Length | 97 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.47 |
| Shannon entropy | 0.46049 |
| G+C content | 0.45115 |
| Mean single sequence MFE | -28.22 |
| Consensus MFE | -16.75 |
| Energy contribution | -18.20 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16623349 97 - 23011544 GGAGAUAAAUCAAAUGCAAAGUUUACUUGUUGACUUUGCGGAC--GGAUGG--------GGCGGUGGUUUGA-GGUUGAGGUUU-GCUCCUCAGCAGAUUUAAAUGCCA ........(((...(((((((((........)))))))))...--.)))..--------((((.........-.(((((((...-...))))))).........)))). ( -29.01, z-score = -1.63, R) >droSim1.chr2L 16319045 97 - 22036055 AGAGAUAAAUCAAAUGCAAAGUUUACUUGUUGACUUUGCGGAC--GGAUGG--------GGCGGUGGUUUGA-GGUUGAGGUUU-GCUCCUCAGCAGAUUUAAAUGCCA ........(((...(((((((((........)))))))))...--.)))..--------((((.........-.(((((((...-...))))))).........)))). ( -29.01, z-score = -1.68, R) >droSec1.super_7 270602 97 - 3727775 AGAGAUAAAUCAAAUGCAAAGUUUACUUGUUGACUUUGCGGAC--GGAUGGGGCGGUGGUUUGA---------GGUUGAGGUUU-GCUCCUCAACGGAUUUAAAUGCCA ........(((...(((((((((........)))))))))...--.)))..((((..(((((..---------.(((((((...-...))))))))))))....)))). ( -28.60, z-score = -1.77, R) >droYak2.chr2R 16658986 96 + 21139217 AGAGAUAAAUCAAAUGCAAAGUUUACUUGUUGACUUUGCGGA---CGGCAG--------GGCGGUGGUUUGA-GGUUGAGGUUU-GCUUCUCAGCAGAUUUAAAUGCCA .........((((((((((((((........))))))))..(---(.((..--------.)).)).))))))-(((..((((((-(((....)))))))))....))). ( -27.60, z-score = -1.30, R) >droEre2.scaffold_4845 19694143 105 - 22589142 GGAGAUAAAUCAAAUGCAAAGUUUACUUGUUGACUUUGCGGAC--GGUCGGGGAGGGGAGGGGGUGGUUUGA-GGUUGAGGUUU-GCUGCUCAGCAGAUUUAAAUGCCA .....(((((((..(.(....((..(((.((((((........--)))))).)))..))).)..))))))).-(((..((((((-(((....)))))))))....))). ( -26.80, z-score = -0.54, R) >droAna3.scaffold_12916 243299 91 + 16180835 GGAGAUAAAUCAAAUGCAAAGUUUACUUGUUGACUUUGCGGACUGGG------------CUUGGUGGUU------UCUGGGUUCGGAGAAGCUUUAGCUUUAAAUACCA ...............((((((((........))))))))((.(((((------------((..(.....------.)..))))))).(((((....))))).....)). ( -27.70, z-score = -2.52, R) >dp4.chr4_group3 8361465 109 - 11692001 AGAGAUAAAUCAAAUGCAAAGUUUACUUGUUGACUUUGUGGGUUACAGUGGGGGGGCGGCUGGGUGGCUGGGCAGUUGAGGUUUCGUCUGGCAAAAGCUUUAAAUGCCA .(((((...((((.(((..((((..((.((((.(((..(.(....).)..)))...)))).))..))))..))).)))).)))))....((((...........)))). ( -28.80, z-score = -0.01, R) >consensus AGAGAUAAAUCAAAUGCAAAGUUUACUUGUUGACUUUGCGGAC__GGAUGG________GGCGGUGGUUUGA_GGUUGAGGUUU_GCUCCUCAGCAGAUUUAAAUGCCA .....((((((...(((((((((........)))))))))..................................(((((((.......))))))).))))))....... (-16.75 = -18.20 + 1.45)

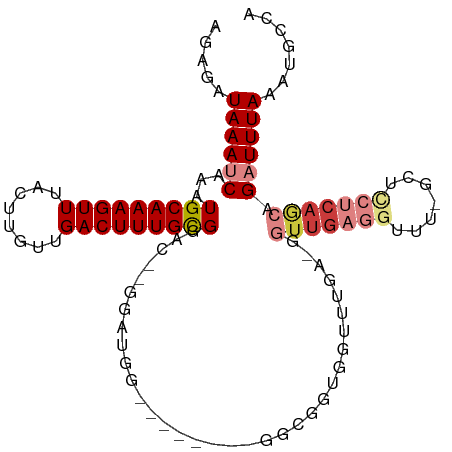
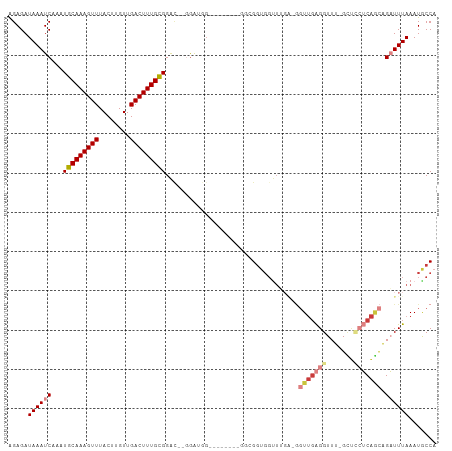
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:43 2011