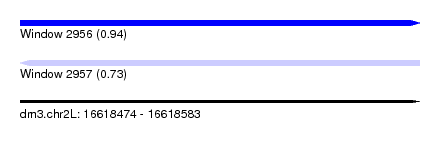
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,618,474 – 16,618,583 |
| Length | 109 |
| Max. P | 0.940074 |

| Location | 16,618,474 – 16,618,583 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 125 |
| Reading direction | forward |
| Mean pairwise identity | 66.59 |
| Shannon entropy | 0.67774 |
| G+C content | 0.56688 |
| Mean single sequence MFE | -28.48 |
| Consensus MFE | -13.99 |
| Energy contribution | -14.67 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.940074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
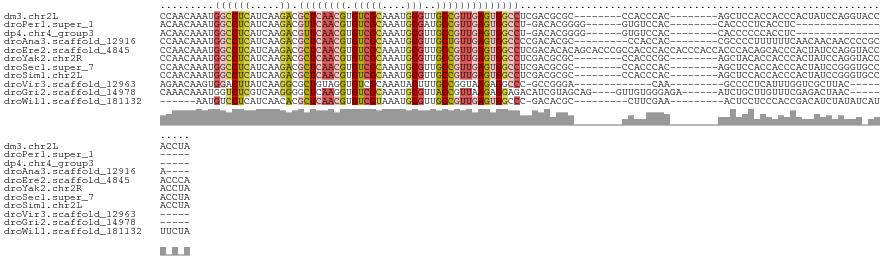
>dm3.chr2L 16618474 109 + 23011544 CCAACAAAUGGCCUCAUCAAGACGCUCAACGUGUCGCAAAUGCGUUGCCGUUGAGUGGCCUCGACGCGC--------CCACCCAC--------AGCUCCACCACCCACUAUCCAGGUACCACCUA .........(((..(.((.((.(((((((((.(((((....)))..)))))))))))..)).)).).))--------).......--------....................(((.....))). ( -25.90, z-score = -0.71, R) >droPer1.super_1 9804474 91 + 10282868 ACAACAAAUGGCCUCAUCAAGACGUUCAACGUGUCGCAAAUGCGAUGCCGUUGAGUGGCCU-GACACGGGG------GUGUCCAC--------CACCCCUCACCUC------------------- .........((((((.....))..(((((((((((((....)))))).))))))).)))).-.....((((------(((.....--------)))))))......------------------- ( -32.90, z-score = -1.95, R) >dp4.chr4_group3 8356121 91 + 11692001 ACAACAAAUGGCCUCAUCAAGACGUUCAACGUGUCGCAAAUGCGUUGCCGUUGAGUGGCCU-GACACGGGG------GUGUCCAC--------CACCCCCCACCUC------------------- .........((((...(((..(((..((((((((.....)))))))).))))))..)))).-.....((((------(((.....--------)))))))......------------------- ( -32.90, z-score = -1.72, R) >droAna3.scaffold_12916 237823 104 - 16180835 CCAACAAAUGGCCUCAUCAAGACGCUCAACGUGUCGCAAAUGCGUUGCUGUUGAGUGGCCCCGACACGC---------CCACCAC--------CGCCCCUUUUUUCAACAACAACCCCGCA---- ..........((........(((((.....)))))((....))((((.(((((((((.......)))((---------.......--------.))........)))))).))))...)).---- ( -22.30, z-score = -0.46, R) >droEre2.scaffold_4845 19689304 125 + 22589142 CCAACAAAUGGCCUCAUCAAGACGCUCAACGUGUCGCAAAUGCGUUGCCGUUGAGUGGCCUCGACACACAGCACCCGCCACCCACCACCCACCACCCACAGCACCCACUAUCCAGGUACCACCCA ........(((...........(((((((((.(((((....)))..)))))))))))((((.((......((....)).....................((......)).)).)))).))).... ( -24.60, z-score = -0.11, R) >droYak2.chr2R 16654119 109 - 21139217 CCAACAAAUGGCCUCAUCAAGACGCUCAACGUGUCGCAAAUGCGUUGCCGUUGAGUGGCCUCGACGCGC--------CCACCCGC--------AGCUACACCACCCACUAUCCAGGUACCACCUA ........((((....((.((.(((((((((.(((((....)))..)))))))))))..)).)).(((.--------.....)))--------.))))...............(((.....))). ( -28.30, z-score = -0.98, R) >droSec1.super_7 265889 109 + 3727775 CCAACAAAUGGCCUCAUCAAGACGCUCAACGUGUCGCAAAUGCGUUGCCGUUGAGUGGCCUCGACGCGC--------CCACCCAC--------AGCUCCACCACCCACUAUCCGGGUGCCACCUA .........((((((.....)).((((((((.(((((....)))..)))))))))))))).....((((--------((......--------....................))))))...... ( -30.97, z-score = -1.14, R) >droSim1.chr2L 16314391 109 + 22036055 CCAACAAAUGGCCUCAUCAAGACGCUCAACGUGUCGCAAAUGCGUUGCCGUUGAGUGGCCUCGACGCGC--------CCACCCAC--------AGCUCCACCACCCACUAUCCGGGUGCCACCUA .........((((((.....)).((((((((.(((((....)))..)))))))))))))).....((((--------((......--------....................))))))...... ( -30.97, z-score = -1.14, R) >droVir3.scaffold_12963 7294835 92 - 20206255 AGAACAAGUGGACUUAUCAAGGCGCUGUAGGUGUCGCAAAUACUUUGCCGGUAAGAGGCCC-GCCGGGA-------------CAA---------GCCCCUCAUUUGGUCGCUUAC---------- .((.(((((((.((((((..(((((.....)))))((((.....)))).)))))).((...-.))(((.-------------...---------..)))))))))).))......---------- ( -27.80, z-score = -0.10, R) >droGri2.scaffold_14978 774993 105 + 1124632 CAAACAAAUGGUCUCGUCAAGGGGCUCAAGGUGUCGCAAAUGCGUUACCGUUAAGAGGAGACAUCGUAGCAG----GUUGUGGGAGA------AUCUGCUUGUUUCGAGACUAAC---------- ........((((((((.((((.(((((..((((.(((....))).))))((((.(((....).)).))))..----.......))).------..)).))))...))))))))..---------- ( -35.60, z-score = -3.11, R) >droWil1.scaffold_181132 965321 100 + 1035393 ------AAUGUCCUCAUCAACACGCUCAACGUGUCGUAAAUGCGUUGCCGUUGAGUGGCCC-GACACGC---------CUUCGAA---------ACUCCUCCCACCGACAUCUAUAUCAUUUCUA ------.(((((..........(((((((((.(((((....)))..)))))))))))...(-((.....---------..)))..---------............))))).............. ( -21.00, z-score = -1.63, R) >consensus CCAACAAAUGGCCUCAUCAAGACGCUCAACGUGUCGCAAAUGCGUUGCCGUUGAGUGGCCUCGACGCGC________CCACCCAC________AGCUCCACCACCCACUAUCCAGGU_CCA_C_A .........((((((.....)).((((((((.(((((....)))..))))))))))))))................................................................. (-13.99 = -14.67 + 0.68)
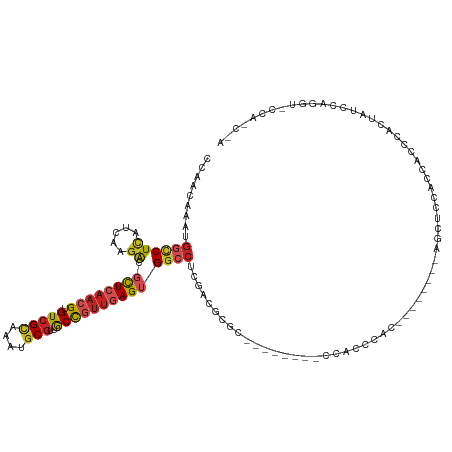
| Location | 16,618,474 – 16,618,583 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 125 |
| Reading direction | reverse |
| Mean pairwise identity | 66.59 |
| Shannon entropy | 0.67774 |
| G+C content | 0.56688 |
| Mean single sequence MFE | -37.26 |
| Consensus MFE | -14.15 |
| Energy contribution | -15.03 |
| Covariance contribution | 0.87 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16618474 109 - 23011544 UAGGUGGUACCUGGAUAGUGGGUGGUGGAGCU--------GUGGGUGG--------GCGCGUCGAGGCCACUCAACGGCAACGCAUUUGCGACACGUUGAGCGUCUUGAUGAGGCCAUUUGUUGG ..(((..((((............))))..)))--------(..(((((--------.(.(((((((((..(((((((....(((....)))...))))))).))))))))).).)))))..)... ( -42.00, z-score = -1.51, R) >droPer1.super_1 9804474 91 - 10282868 -------------------GAGGUGAGGGGUG--------GUGGACAC------CCCCGUGUC-AGGCCACUCAACGGCAUCGCAUUUGCGACACGUUGAACGUCUUGAUGAGGCCAUUUGUUGU -------------------((..((.((((((--------.....)))------)))))..))-.((((..((((((...((((....))))..)))))).(((....))).))))......... ( -37.40, z-score = -1.85, R) >dp4.chr4_group3 8356121 91 - 11692001 -------------------GAGGUGGGGGGUG--------GUGGACAC------CCCCGUGUC-AGGCCACUCAACGGCAACGCAUUUGCGACACGUUGAACGUCUUGAUGAGGCCAUUUGUUGU -------------------..(..((((((((--------.....)))------)))).)..)-.((((..((((((....(((....)))...)))))).(((....))).))))......... ( -36.70, z-score = -1.28, R) >droAna3.scaffold_12916 237823 104 + 16180835 ----UGCGGGGUUGUUGUUGAAAAAAGGGGCG--------GUGGUGG---------GCGUGUCGGGGCCACUCAACAGCAACGCAUUUGCGACACGUUGAGCGUCUUGAUGAGGCCAUUUGUUGG ----((((..(((((((((....)).......--------..(((((---------.(.......).))))))))))))..))))....(((((...((.((.((.....)).))))..))))). ( -32.40, z-score = 0.31, R) >droEre2.scaffold_4845 19689304 125 - 22589142 UGGGUGGUACCUGGAUAGUGGGUGCUGUGGGUGGUGGGUGGUGGGUGGCGGGUGCUGUGUGUCGAGGCCACUCAACGGCAACGCAUUUGCGACACGUUGAGCGUCUUGAUGAGGCCAUUUGUUGG ...(..((((((.......))))))..)....((..(((((((.(..((....))..).)((((((((..(((((((....(((....)))...))))))).))))))))...))))))..)).. ( -44.90, z-score = -0.41, R) >droYak2.chr2R 16654119 109 + 21139217 UAGGUGGUACCUGGAUAGUGGGUGGUGUAGCU--------GCGGGUGG--------GCGCGUCGAGGCCACUCAACGGCAACGCAUUUGCGACACGUUGAGCGUCUUGAUGAGGCCAUUUGUUGG ..(((..((((............))))..)))--------((((((((--------.(.(((((((((..(((((((....(((....)))...))))))).))))))))).).))))))))... ( -43.80, z-score = -1.76, R) >droSec1.super_7 265889 109 - 3727775 UAGGUGGCACCCGGAUAGUGGGUGGUGGAGCU--------GUGGGUGG--------GCGCGUCGAGGCCACUCAACGGCAACGCAUUUGCGACACGUUGAGCGUCUUGAUGAGGCCAUUUGUUGG .......((((((.....))))))......(.--------(..(((((--------.(.(((((((((..(((((((....(((....)))...))))))).))))))))).).)))))..).). ( -45.20, z-score = -1.79, R) >droSim1.chr2L 16314391 109 - 22036055 UAGGUGGCACCCGGAUAGUGGGUGGUGGAGCU--------GUGGGUGG--------GCGCGUCGAGGCCACUCAACGGCAACGCAUUUGCGACACGUUGAGCGUCUUGAUGAGGCCAUUUGUUGG .......((((((.....))))))......(.--------(..(((((--------.(.(((((((((..(((((((....(((....)))...))))))).))))))))).).)))))..).). ( -45.20, z-score = -1.79, R) >droVir3.scaffold_12963 7294835 92 + 20206255 ----------GUAAGCGACCAAAUGAGGGGC---------UUG-------------UCCCGGC-GGGCCUCUUACCGGCAAAGUAUUUGCGACACCUACAGCGCCUUGAUAAGUCCACUUGUUCU ----------...........((..((((((---------(((-------------((..(((-(.(((.......)))...(((..((...))..)))..))))..))))))))).))..)).. ( -28.10, z-score = -0.57, R) >droGri2.scaffold_14978 774993 105 - 1124632 ----------GUUAGUCUCGAAACAAGCAGAU------UCUCCCACAAC----CUGCUACGAUGUCUCCUCUUAACGGUAACGCAUUUGCGACACCUUGAGCCCCUUGACGAGACCAUUUGUUUG ----------....(((((......(((((..------...........----))))).....(((....(((((.(((..(((....)))..))))))))......)))))))).......... ( -24.92, z-score = -2.44, R) >droWil1.scaffold_181132 965321 100 - 1035393 UAGAAAUGAUAUAGAUGUCGGUGGGAGGAGU---------UUCGAAG---------GCGUGUC-GGGCCACUCAACGGCAACGCAUUUACGACACGUUGAGCGUGUUGAUGAGGACAUU------ .............(((((((((..((.(.((---------(.....)---------)).).))-..))).((((.((....))......(((((((.....))))))).))))))))))------ ( -29.20, z-score = -1.28, R) >consensus U_G_UGG_ACCUGGAUAGUGGGUGGAGGAGCU________GUGGGUGG________GCGCGUCGAGGCCACUCAACGGCAACGCAUUUGCGACACGUUGAGCGUCUUGAUGAGGCCAUUUGUUGG ...........................................................((((.((((..(((((((....(((....)))...))))))).)))).)))).............. (-14.15 = -15.03 + 0.87)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:42 2011