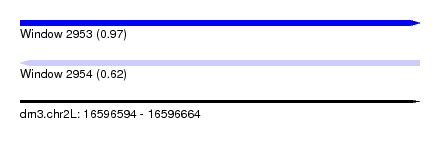
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,596,594 – 16,596,664 |
| Length | 70 |
| Max. P | 0.969963 |

| Location | 16,596,594 – 16,596,664 |
|---|---|
| Length | 70 |
| Sequences | 7 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 80.46 |
| Shannon entropy | 0.38440 |
| G+C content | 0.42107 |
| Mean single sequence MFE | -12.83 |
| Consensus MFE | -7.79 |
| Energy contribution | -8.91 |
| Covariance contribution | 1.13 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969963 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
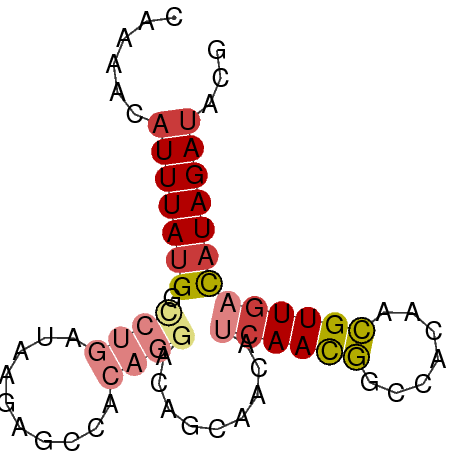
>dm3.chr2L 16596594 70 + 23011544 CGAAACAUUUAUGGCCUGAUAAGAGCCACAGGACAGCAACAUCAACGGCCACAACGUUGACAUAGAUACG ......(((((((.((((..........)))).........((((((.......)))))))))))))... ( -15.40, z-score = -3.12, R) >droEre2.scaffold_4845 19664741 70 + 22589142 CAAAACAUUUAUGGCCUGAUAGGAGCCACAGGACAACAAAAUCAACGGCCACAACGUUGACAUAGAUACG ......(((((((.((((..........)))).........((((((.......)))))))))))))... ( -15.40, z-score = -3.37, R) >droYak2.chr2R 16632023 70 - 21139217 CAAAACAUUUAUGGUCUGAUAAGAGCCACAGGACAGCAACAUCAACGGCCACAACGUUGACAUAGAUACG ......(((((((((((.............)))).......((((((.......)))))))))))))... ( -14.42, z-score = -2.64, R) >droSec1.super_7 244071 70 + 3727775 CAAAACAUUUAUGGCCUGAUAAGAGCCACAGGACAGCAACAUCAACGGCCACAACGUUGACAUAGAUACG ......(((((((.((((..........)))).........((((((.......)))))))))))))... ( -15.40, z-score = -3.31, R) >droSim1.chr2L 16292145 70 + 22036055 CAAAACAUUUAUGGCCUGAUAAGAGCCACAGGACAGCAACAUCAACGGCCACAACGUUGACAUAGAUACG ......(((((((.((((..........)))).........((((((.......)))))))))))))... ( -15.40, z-score = -3.31, R) >dp4.chr4_group3 8334751 64 + 11692001 AAGCACAUUUAUGGCCUGAUAACACUCA------AACAACAACAACAACAACCAUGUUGACAUAGAUAUG ...((.(((((((...(((......)))------...........(((((....))))).))))))).)) ( -8.70, z-score = -1.73, R) >droAna3.scaffold_12916 215256 60 - 16180835 CAAAACAUUUAUGGCCCGAUAACGUUUA------AACAACA--AAUA--CUUGGUGUUGAUUUAGAAAUG ........................((((------(((((((--....--.....))))).)))))).... ( -5.10, z-score = 0.85, R) >consensus CAAAACAUUUAUGGCCUGAUAAGAGCCACAGGACAGCAACAUCAACGGCCACAACGUUGACAUAGAUACG ......(((((((.((((..........)))).........((((((.......)))))))))))))... ( -7.79 = -8.91 + 1.13)

| Location | 16,596,594 – 16,596,664 |
|---|---|
| Length | 70 |
| Sequences | 7 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 80.46 |
| Shannon entropy | 0.38440 |
| G+C content | 0.42107 |
| Mean single sequence MFE | -15.20 |
| Consensus MFE | -9.71 |
| Energy contribution | -10.56 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.622189 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 16596594 70 - 23011544 CGUAUCUAUGUCAACGUUGUGGCCGUUGAUGUUGCUGUCCUGUGGCUCUUAUCAGGCCAUAAAUGUUUCG ............((((((((((((..(((((..(((.......)))...))))))))))).))))))... ( -19.40, z-score = -2.50, R) >droEre2.scaffold_4845 19664741 70 - 22589142 CGUAUCUAUGUCAACGUUGUGGCCGUUGAUUUUGUUGUCCUGUGGCUCCUAUCAGGCCAUAAAUGUUUUG ............((((((((((((..((((...(..(((....)))..).)))))))))).))))))... ( -16.80, z-score = -2.08, R) >droYak2.chr2R 16632023 70 + 21139217 CGUAUCUAUGUCAACGUUGUGGCCGUUGAUGUUGCUGUCCUGUGGCUCUUAUCAGACCAUAAAUGUUUUG ............((((((((((...((((((..(((.......)))...)))))).)))).))))))... ( -14.30, z-score = -0.79, R) >droSec1.super_7 244071 70 - 3727775 CGUAUCUAUGUCAACGUUGUGGCCGUUGAUGUUGCUGUCCUGUGGCUCUUAUCAGGCCAUAAAUGUUUUG ............((((((((((((..(((((..(((.......)))...))))))))))).))))))... ( -19.40, z-score = -2.55, R) >droSim1.chr2L 16292145 70 - 22036055 CGUAUCUAUGUCAACGUUGUGGCCGUUGAUGUUGCUGUCCUGUGGCUCUUAUCAGGCCAUAAAUGUUUUG ............((((((((((((..(((((..(((.......)))...))))))))))).))))))... ( -19.40, z-score = -2.55, R) >dp4.chr4_group3 8334751 64 - 11692001 CAUAUCUAUGUCAACAUGGUUGUUGUUGUUGUUGUU------UGAGUGUUAUCAGGCCAUAAAUGUGCUU (((((.((((.(((((....)))))........(((------(((......)))))))))).)))))... ( -13.20, z-score = -0.83, R) >droAna3.scaffold_12916 215256 60 + 16180835 CAUUUCUAAAUCAACACCAAG--UAUU--UGUUGUU------UAAACGUUAUCGGGCCAUAAAUGUUUUG ................((..(--((..--((((...------..)))).)))..)).............. ( -3.90, z-score = 1.21, R) >consensus CGUAUCUAUGUCAACGUUGUGGCCGUUGAUGUUGCUGUCCUGUGGCUCUUAUCAGGCCAUAAAUGUUUUG ............((((((((((((..(((((..(((.......)))...))))))))))).))))))... ( -9.71 = -10.56 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:39 2011