| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,570,348 – 16,570,439 |
| Length | 91 |
| Max. P | 0.888669 |

| Location | 16,570,348 – 16,570,439 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 77.84 |
| Shannon entropy | 0.40497 |
| G+C content | 0.50736 |
| Mean single sequence MFE | -22.46 |
| Consensus MFE | -12.28 |
| Energy contribution | -13.75 |
| Covariance contribution | 1.47 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584686 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
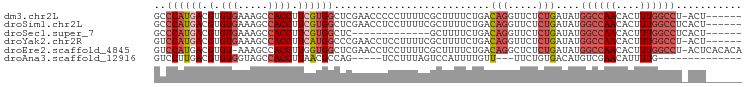
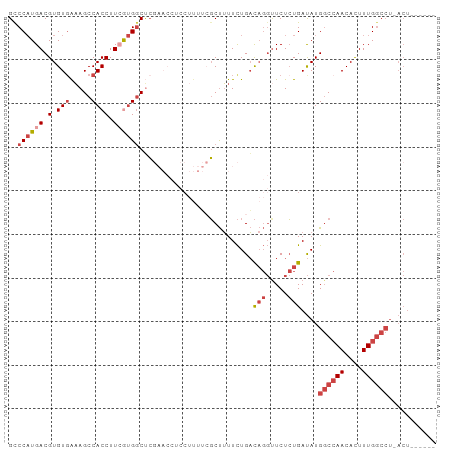
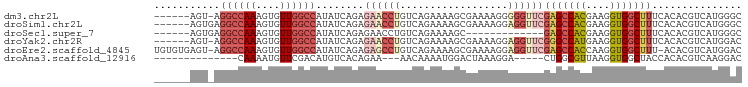
>dm3.chr2L 16570348 91 + 23011544 GCCCAUGACGUGUGAAAGCCACCUUCGUGGCUCGAACCCCCUUUUCGCUUUUCUGACAGGUUCUCUGAUAUGGCCAACACUUUGGCCU-ACU------ (((.((((.(.(((.....)))).)))))))..(((((....................)))))........((((((....)))))).-...------ ( -22.85, z-score = -1.43, R) >droSim1.chr2L 16264845 92 + 22036055 GCCCAUGACGUGUGAAAGCCACCUUCGUGGCUCGAACCUCCUUUUCGCUUUUCUGACAGGUUCUCUGAUAUGGCCAACACUUUGGCCUCACU------ ...........((((.((((((....)))))).((((((.....(((......))).))))))........((((((....)))))))))).------ ( -26.00, z-score = -2.30, R) >droSec1.super_7 217521 79 + 3727775 GCCCAUGACGUGUGAAAGCCACCUUCGUGGCUC-------------GCUUUUCUGACAGGUUCUCUGAUAUGGCCAACACUUUGGCCUCACU------ ...........((((.((((((....)))))).-------------..........(((.....)))....((((((....)))))))))).------ ( -21.80, z-score = -1.18, R) >droYak2.chr2R 16606284 91 - 21139217 GUCCAUGACGUGUGAAAGCCACCUUCAUGGCCCGAACCUCCUUUUCGCUUUUCUGACAGGUUCUCUGAUAUGGCCAACACUUUGGCCU-ACU------ ..((((((.(.(((.....)))).))))))...((((((.....(((......))).))))))........((((((....)))))).-...------ ( -24.50, z-score = -2.28, R) >droEre2.scaffold_4845 19638610 96 + 22589142 GUCCAUGACGUGU-AAAGCCACCUUGGUGGCUCGAACCUCCUUUUCGCUUUUCUGACAGGCUCUCUGAUAUGGCCAACACUUUGGCCU-ACUCACACA (((...)))((((-..((((((....))))))..............((((.......)))).....((...((((((....)))))).-..)))))). ( -26.40, z-score = -2.16, R) >droAna3.scaffold_12916 189088 76 - 16180835 GUCCUUGACGUGUGGUAGCCACCUUAACGCCAG-----UCCUUUAGUCCAUUUUGUU---UUCUGUGACAUGUCGAACAUUUUG-------------- ....((((((((((((....)))....(((.((-----.....(((......)))..---..))))))))))))))........-------------- ( -13.20, z-score = -0.45, R) >consensus GCCCAUGACGUGUGAAAGCCACCUUCGUGGCUCGAACCUCCUUUUCGCUUUUCUGACAGGUUCUCUGAUAUGGCCAACACUUUGGCCU_ACU______ ..((((((.(.(((.....)))).))))))..........................(((.....)))....((((((....))))))........... (-12.28 = -13.75 + 1.47)
| Location | 16,570,348 – 16,570,439 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 77.84 |
| Shannon entropy | 0.40497 |
| G+C content | 0.50736 |
| Mean single sequence MFE | -27.75 |
| Consensus MFE | -15.16 |
| Energy contribution | -17.44 |
| Covariance contribution | 2.28 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.888669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
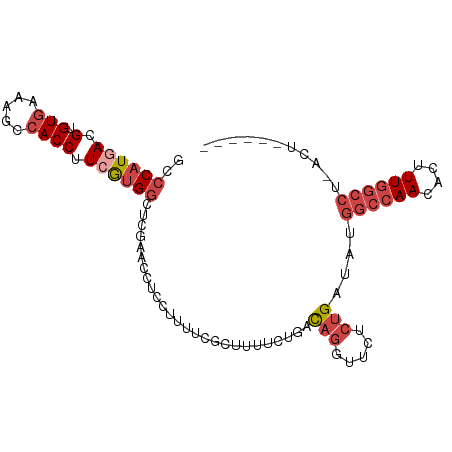
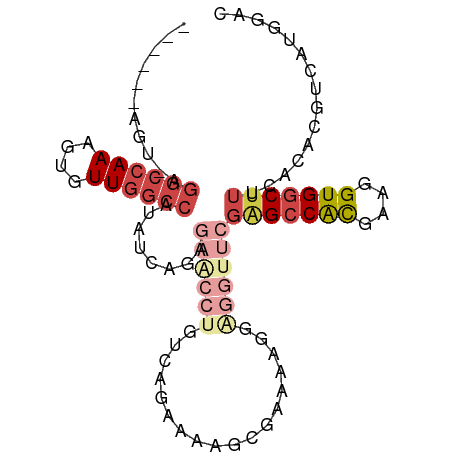
>dm3.chr2L 16570348 91 - 23011544 ------AGU-AGGCCAAAGUGUUGGCCAUAUCAGAGAACCUGUCAGAAAAGCGAAAAGGGGGUUCGAGCCACGAAGGUGGCUUUCACACGUCAUGGGC ------...-.((((((....))))))...((((((..(((.((........))...)))..)))(((((((....)))))))..........))).. ( -30.40, z-score = -2.19, R) >droSim1.chr2L 16264845 92 - 22036055 ------AGUGAGGCCAAAGUGUUGGCCAUAUCAGAGAACCUGUCAGAAAAGCGAAAAGGAGGUUCGAGCCACGAAGGUGGCUUUCACACGUCAUGGGC ------.((((((((((....))))))........((((((.((........)).....))))))(((((((....)))))))))))........... ( -33.30, z-score = -3.08, R) >droSec1.super_7 217521 79 - 3727775 ------AGUGAGGCCAAAGUGUUGGCCAUAUCAGAGAACCUGUCAGAAAAGC-------------GAGCCACGAAGGUGGCUUUCACACGUCAUGGGC ------.((((((((((....))))))....(((.....)))..........-------------(((((((....)))))))))))........... ( -27.30, z-score = -2.17, R) >droYak2.chr2R 16606284 91 + 21139217 ------AGU-AGGCCAAAGUGUUGGCCAUAUCAGAGAACCUGUCAGAAAAGCGAAAAGGAGGUUCGGGCCAUGAAGGUGGCUUUCACACGUCAUGGAC ------...-.((((((....))))))........((((((.((........)).....))))))...((((((..(((.......))).)))))).. ( -27.90, z-score = -1.71, R) >droEre2.scaffold_4845 19638610 96 - 22589142 UGUGUGAGU-AGGCCAAAGUGUUGGCCAUAUCAGAGAGCCUGUCAGAAAAGCGAAAAGGAGGUUCGAGCCACCAAGGUGGCUUU-ACACGUCAUGGAC .((((((..-.((((((....))))))........((((((.((........)).....)))))).((((((....))))))))-))))......... ( -32.80, z-score = -2.82, R) >droAna3.scaffold_12916 189088 76 + 16180835 --------------CAAAAUGUUCGACAUGUCACAGAA---AACAAAAUGGACUAAAGGA-----CUGGCGUUAAGGUGGCUACCACACGUCAAGGAC --------------......((((.....(((.((...---.......)))))....)))-----)((((((....((((...))))))))))..... ( -14.80, z-score = -0.48, R) >consensus ______AGU_AGGCCAAAGUGUUGGCCAUAUCAGAGAACCUGUCAGAAAAGCGAAAAGGAGGUUCGAGCCACGAAGGUGGCUUUCACACGUCAUGGAC ...........((((((....))))))........((((((..................))))))(((((((....)))))))............... (-15.16 = -17.44 + 2.28)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:36 2011