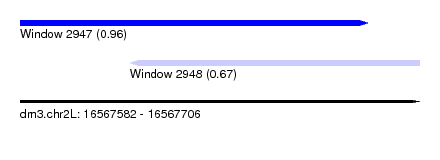
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,567,582 – 16,567,706 |
| Length | 124 |
| Max. P | 0.956331 |

| Location | 16,567,582 – 16,567,690 |
|---|---|
| Length | 108 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 71.31 |
| Shannon entropy | 0.46154 |
| G+C content | 0.30545 |
| Mean single sequence MFE | -23.20 |
| Consensus MFE | -13.19 |
| Energy contribution | -12.70 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.63 |
| SVM RNA-class probability | 0.956331 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
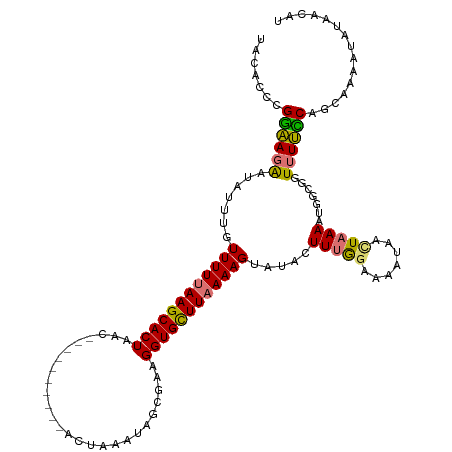
>dm3.chr2L 16567582 108 + 23011544 UACACCCGGAAGAAUAUUUGUUUUUAAGCACUAA-----------AGUAAAUAGCGAAGGUGCUUAAAAGUAAACUUUA-AAAACAACUAAAAUGGUGGUUUUCCAGCAAAAUAUAACAU ..(((((....)....((((((((((((((((..-----------.((.....))...)))))))))))))))).....-..............))))...................... ( -21.60, z-score = -2.06, R) >droSim1.chr2L 16262031 120 + 22036055 UACACCCGGAAGAAUAUUUGUUUUUAAGCACUAACUGCGAUUUAAAGUAAAUAGCGAAGGUGCUUAAAAGUUUGCUUUGGAAAAUAACUAAAAUGGCGGUUUUCCAGCAAAAUAUAACAU ......(....)........((((((((((((...(((.(((((...))))).)))..))))))))))))((((((..(((((((..((.....))..)))))))))))))......... ( -29.30, z-score = -2.60, R) >droSec1.super_7 214713 110 + 3727775 UACACCCGGAAGAAUAUUUGUUUUUAAGCACUAAC---------GGCGAUUUACCGAAGGUGCUUAAAAGUAUACUUUGGAAAAUAACUAAA-UGGCGGUUUUUCAGCAAAAUACAAAAU ......(....).........(((((((((((..(---------((.......)))..)))))))))))((((...(((((((((..((...-.))..)))))))))....))))..... ( -25.30, z-score = -2.42, R) >droEre2.scaffold_4845 19635961 106 + 22589142 UACACCCGAUAGGACAUUUGUUUUUAAGCACUUACUACGAUUUACACUUUAAUGCAAAGGUGUUUUAAAGUAUAGUUAGGAAAAUAAUUAAAGUAA-AUUAUAUCAU------------- .......((((((((....))))...............((((((((((((((.(((....))).))))))).((((((......))))))..))))-))).))))..------------- ( -16.60, z-score = -0.91, R) >consensus UACACCCGGAAGAAUAUUUGUUUUUAAGCACUAAC__________ACUAAAUAGCGAAGGUGCUUAAAAGUAUACUUUGGAAAAUAACUAAAAUGGCGGUUUUCCAGCAAAAUAUAACAU .......((((((.......((((((((((((..........................)))))))))))).....(((((.......))))).......))))))............... (-13.19 = -12.70 + -0.50)

| Location | 16,567,616 – 16,567,706 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 79.28 |
| Shannon entropy | 0.28285 |
| G+C content | 0.31756 |
| Mean single sequence MFE | -16.64 |
| Consensus MFE | -15.57 |
| Energy contribution | -15.69 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.18 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.667681 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
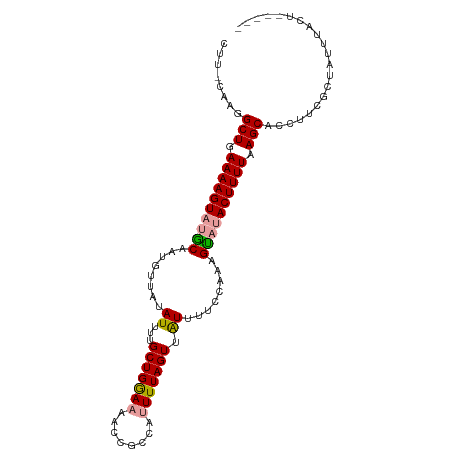
>dm3.chr2L 16567616 90 - 23011544 -------GGCUGAAAAGUAUACAAUGUUAUAUUUUGCUGGAAAACCACCAUUUUAGUUGUUUU-UAAAGUUUACUUUUAAGCACCUUCGCUAUUUACU----- -------.(((.(((((((.((....(((......(((((((........)))))))......-))).)).))))))).)))................----- ( -14.10, z-score = -0.43, R) >droSim1.chr2L 16262071 103 - 22036055 CUUCCAAGGCUGAAAAGUAUGCAAUGUUAUAUUUUGCUGGAAAACCGCCAUUUUAGUUAUUUUCCAAAGCAAACUUUUAAGCACCUUCGCUAUUUACUUUAAA ........(((.((((((.(((...((........))(((((((..((.......))..)))))))..))).)))))).)))..................... ( -18.50, z-score = -0.86, R) >droSec1.super_7 214753 93 - 3727775 CUUUCAAGGCUGAAAAGUAUGCAAUUUUGUAUUUUGCUGAAAAACCGCCA-UUUAGUUAUUUUCCAAAGUAUACUUUUAAGCACCUUCGGUAAA--------- ........(((.((((((((((...((((......((((((.........-)))))).......)))))))))))))).)))(((...)))...--------- ( -17.32, z-score = -1.12, R) >consensus CUU_CAAGGCUGAAAAGUAUGCAAUGUUAUAUUUUGCUGGAAAACCGCCAUUUUAGUUAUUUUCCAAAGUAUACUUUUAAGCACCUUCGCUAUUUACU_____ ........(((.((((((((((........((...(((((((........))))))).))........)))))))))).)))..................... (-15.57 = -15.69 + 0.12)

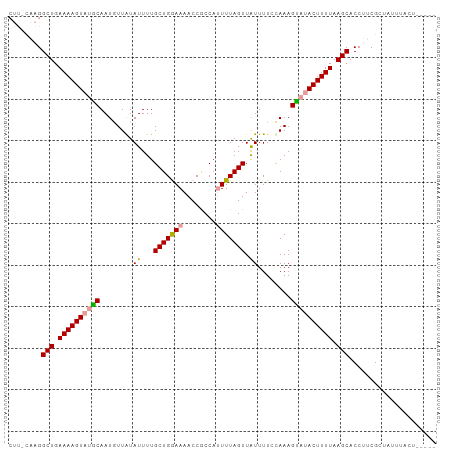
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:34 2011