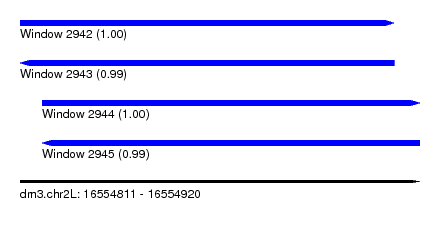
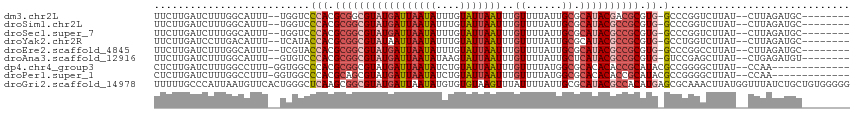
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,554,811 – 16,554,920 |
| Length | 109 |
| Max. P | 0.999608 |

| Location | 16,554,811 – 16,554,913 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.43914 |
| G+C content | 0.45666 |
| Mean single sequence MFE | -33.50 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.13 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.35 |
| Mean z-score | -3.38 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.78 |
| SVM RNA-class probability | 0.999304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
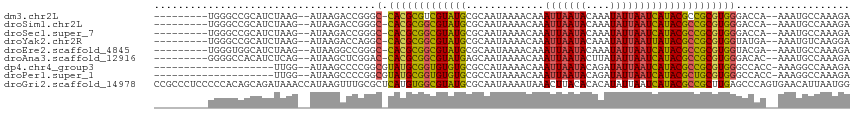
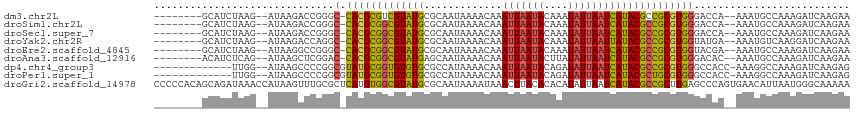
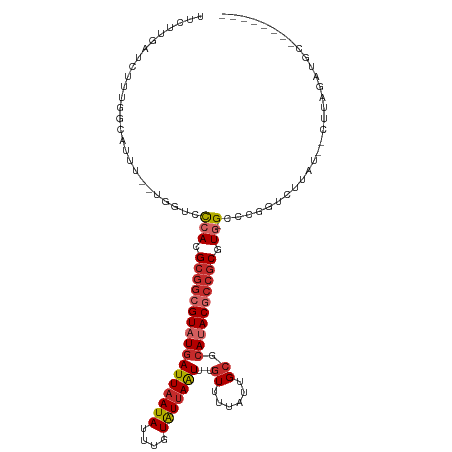
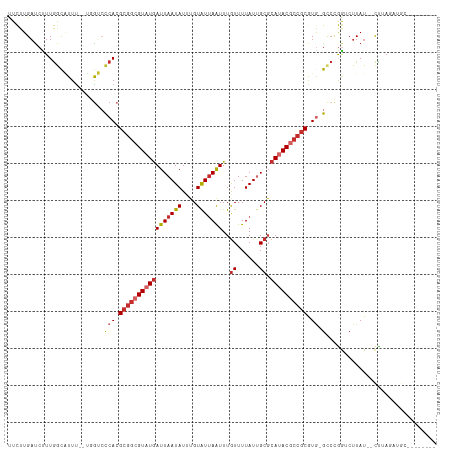
>dm3.chr2L 16554811 102 + 23011544 ---------UGGGCCGCAUCUAAG--AUAAGACCGGGC-CACGCGUCGUAUGCGCAAUAAAACAAAUUAAUACAAAUAUUAAUCAUACGCCGCGUGGGACCA--AAAUGCCAAAGA ---------..(((....(((...--...)))..((.(-((((((.((((((.............(((((((....))))))))))))).)))))))..)).--....)))..... ( -28.91, z-score = -1.98, R) >droSim1.chr2L 16250740 102 + 22036055 ---------UGGGCCGCAUCUAAG--AUAAGACCGGGC-CACGCGGCGUAUGCGCAAUAAAACAAAUUAAUACAAAUAUUAAUCAUACGCCGCGUGGGACCA--AAAUGCCAAAGA ---------..(((....(((...--...)))..((.(-(((((((((((((.............(((((((....)))))))))))))))))))))..)).--....)))..... ( -36.01, z-score = -3.86, R) >droSec1.super_7 201481 102 + 3727775 ---------UGGGCCGCAUCUAAG--AUAAGACCGGGC-CACGCGGCGUAUGCGCAAUAAAACAAAUUAAUACAAAUAUUAAUCAUACGCCGCGUGGGACCA--AAAUGCCAAAGA ---------..(((....(((...--...)))..((.(-(((((((((((((.............(((((((....)))))))))))))))))))))..)).--....)))..... ( -36.01, z-score = -3.86, R) >droYak2.chr2R 16590762 102 - 21139217 ---------UGGGCCGCAUCUAAG--AUAAGACCAGGC-CACGCGGCGUAUGCGCAAUAAAACAAAUUAAUACAAAUAUUAAUUAUACGCCGCGUGGUAUGA--AAAUGUCAAGGA ---------..((((...(((...--...)))...)))-)((((((((((((.............(((((((....)))))))))))))))))))(..((..--..))..)..... ( -32.51, z-score = -3.27, R) >droEre2.scaffold_4845 19623684 102 + 22589142 ---------UGGGUGGCAUCUAAG--AUAAGGCCGGGC-CACGCGGCGUAUGCGCAAUAAAACAAAUUAAUACAAAUAUUAAUCAUACGCCGCGUGGUACGA--AAAUGCCAAAGA ---------....((((((((...--...))..((..(-(((((((((((((.............(((((((....)))))))))))))))))))))..)).--..)))))).... ( -39.11, z-score = -4.47, R) >droAna3.scaffold_12916 175829 102 - 16180835 ---------GGGGCCACAUCUCAG--AUAAGCUCGGAC-CACGCGGCGUAUGAGCAAUAAAACAAAUUAAUACUUAUAUUAAUCAUACGCCGCGUGGGACAC--AAAUGCCAAAGA ---------..(((.........(--(.....))(..(-((((((((((((((..((((.................))))..)))))))))))))))..)..--....)))..... ( -37.23, z-score = -5.87, R) >dp4.chr4_group3 8284209 93 + 11692001 --------------------UUGG--AUAAGCCCCGGCGUAUGCGGUGUGUGCGCCAUAAAACAAAUUAAUACAGAUAUUAAUCAUACGCCGCGUGGGCCACC-AAAGGCCAAAGA --------------------((((--....(((((((((((((.((((....)))).........(((((((....))))))))))))))))...))))..))-)).......... ( -31.30, z-score = -2.05, R) >droPer1.super_1 9733073 93 + 10282868 --------------------UUGG--AUAAGCCCCGGCGUAUGCGGUGUGUGCGCCAUAAAACAAAUUAAUACAGAUAUUAAUCAUACGCUGCGUGGGCCACC-AAAGGCCAAAGA --------------------((((--....(((((((((((((.((((....)))).........(((((((....))))))))))))))))...))))..))-)).......... ( -29.10, z-score = -1.41, R) >droGri2.scaffold_14978 740446 116 + 1124632 CCGCCCUCCCCCACAGCAGAUAAACCAUAAGUUUGCGCUCAUGUGGCGUAUGCGCAAUAAAAUAAACUUACACACAUAUUAAUCAUACGCCGCUUGAGCCCAGUGAACAUUAAUGG ...........(((.((((((.........))))))(((((.((((((((((...((((.................))))...)))))))))).)))))...)))........... ( -31.33, z-score = -3.66, R) >consensus _________UGGGCCGCAUCUAAG__AUAAGACCGGGC_CACGCGGCGUAUGCGCAAUAAAACAAAUUAAUACAAAUAUUAAUCAUACGCCGCGUGGGACCA__AAAUGCCAAAGA .......................................(((((((((((((.............(((((((....)))))))))))))))))))).................... (-21.53 = -21.13 + -0.40)

| Location | 16,554,811 – 16,554,913 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 77.86 |
| Shannon entropy | 0.43914 |
| G+C content | 0.45666 |
| Mean single sequence MFE | -35.60 |
| Consensus MFE | -15.75 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.54 |
| SVM RNA-class probability | 0.992388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16554811 102 - 23011544 UCUUUGGCAUUU--UGGUCCCACGCGGCGUAUGAUUAAUAUUUGUAUUAAUUUGUUUUAUUGCGCAUACGACGCGUG-GCCCGGUCUUAU--CUUAGAUGCGGCCCA--------- .....(((....--(((..(((((((.(((((((((((((....)))))))..((......)).)))))).))))))-).)))((((...--...))))...)))..--------- ( -32.20, z-score = -1.95, R) >droSim1.chr2L 16250740 102 - 22036055 UCUUUGGCAUUU--UGGUCCCACGCGGCGUAUGAUUAAUAUUUGUAUUAAUUUGUUUUAUUGCGCAUACGCCGCGUG-GCCCGGUCUUAU--CUUAGAUGCGGCCCA--------- .....(((....--(((..(((((((((((((((((((((....)))))))..((......)).)))))))))))))-).)))((((...--...))))...)))..--------- ( -39.30, z-score = -3.96, R) >droSec1.super_7 201481 102 - 3727775 UCUUUGGCAUUU--UGGUCCCACGCGGCGUAUGAUUAAUAUUUGUAUUAAUUUGUUUUAUUGCGCAUACGCCGCGUG-GCCCGGUCUUAU--CUUAGAUGCGGCCCA--------- .....(((....--(((..(((((((((((((((((((((....)))))))..((......)).)))))))))))))-).)))((((...--...))))...)))..--------- ( -39.30, z-score = -3.96, R) >droYak2.chr2R 16590762 102 + 21139217 UCCUUGACAUUU--UCAUACCACGCGGCGUAUAAUUAAUAUUUGUAUUAAUUUGUUUUAUUGCGCAUACGCCGCGUG-GCCUGGUCUUAU--CUUAGAUGCGGCCCA--------- ............--.....(((((((((((((((((((((....)))))))).((......))..))))))))))))-)...((((.(((--(...)))).))))..--------- ( -34.00, z-score = -3.87, R) >droEre2.scaffold_4845 19623684 102 - 22589142 UCUUUGGCAUUU--UCGUACCACGCGGCGUAUGAUUAAUAUUUGUAUUAAUUUGUUUUAUUGCGCAUACGCCGCGUG-GCCCGGCCUUAU--CUUAGAUGCCACCCA--------- ....((((((((--(((..(((((((((((((((((((((....)))))))..((......)).)))))))))))))-)..)))......--...))))))))....--------- ( -41.70, z-score = -5.82, R) >droAna3.scaffold_12916 175829 102 + 16180835 UCUUUGGCAUUU--GUGUCCCACGCGGCGUAUGAUUAAUAUAAGUAUUAAUUUGUUUUAUUGCUCAUACGCCGCGUG-GUCCGAGCUUAU--CUGAGAUGUGGCCCC--------- .....(((....--.((..(((((((((((((((.((((((((((....))))))...)))).))))))))))))))-)..))..(((..--..))).....)))..--------- ( -38.40, z-score = -4.77, R) >dp4.chr4_group3 8284209 93 - 11692001 UCUUUGGCCUUU-GGUGGCCCACGCGGCGUAUGAUUAAUAUCUGUAUUAAUUUGUUUUAUGGCGCACACACCGCAUACGCCGGGGCUUAU--CCAA-------------------- ..........((-((.(((((...((((((((((((((((....))))))).........((.(....).)).))))))))))))))...--))))-------------------- ( -30.30, z-score = -1.56, R) >droPer1.super_1 9733073 93 - 10282868 UCUUUGGCCUUU-GGUGGCCCACGCAGCGUAUGAUUAAUAUCUGUAUUAAUUUGUUUUAUGGCGCACACACCGCAUACGCCGGGGCUUAU--CCAA-------------------- ..........((-((.(((((.((..((((((((((((((....))))))).........((.(....).)).))))))))))))))...--))))-------------------- ( -26.90, z-score = -0.85, R) >droGri2.scaffold_14978 740446 116 - 1124632 CCAUUAAUGUUCACUGGGCUCAAGCGGCGUAUGAUUAAUAUGUGUGUAAGUUUAUUUUAUUGCGCAUACGCCACAUGAGCGCAAACUUAUGGUUUAUCUGCUGUGGGGGAGGGCGG .......(((((..((.(((((.(.((((((((..(((((...(((......)))..)))))..)))))))).).))))).))..((((((((......))))))))...))))). ( -38.30, z-score = -2.10, R) >consensus UCUUUGGCAUUU__UGGUCCCACGCGGCGUAUGAUUAAUAUUUGUAUUAAUUUGUUUUAUUGCGCAUACGCCGCGUG_GCCCGGUCUUAU__CUUAGAUGCGGCCCA_________ ....(((.((......)).))).(((((((((((((((((....)))))))..((......)).)))))))))).......................................... (-15.75 = -16.43 + 0.68)
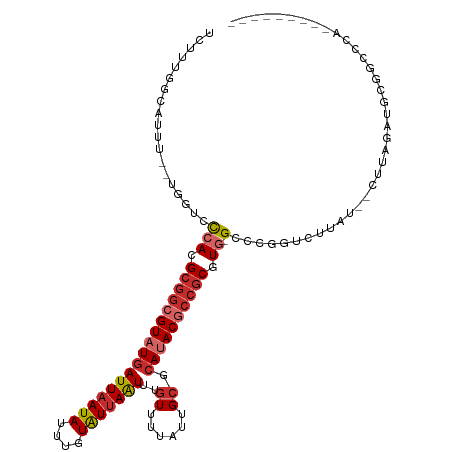
| Location | 16,554,817 – 16,554,920 |
|---|---|
| Length | 103 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.88 |
| Shannon entropy | 0.37926 |
| G+C content | 0.42973 |
| Mean single sequence MFE | -32.43 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.13 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.35 |
| Mean z-score | -4.07 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999608 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
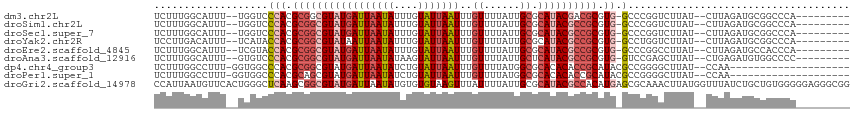
>dm3.chr2L 16554817 103 + 23011544 --------GCAUCUAAG--AUAAGACCGGGC-CACGCGUCGUAUGCGCAAUAAAACAAAUUAAUACAAAUAUUAAUCAUACGCCGCGUGGGACCA--AAAUGCCAAAGAUCAAGAA --------((((((...--...)))..((.(-((((((.((((((.............(((((((....))))))))))))).)))))))..)).--...)))............. ( -26.41, z-score = -2.87, R) >droSim1.chr2L 16250746 103 + 22036055 --------GCAUCUAAG--AUAAGACCGGGC-CACGCGGCGUAUGCGCAAUAAAACAAAUUAAUACAAAUAUUAAUCAUACGCCGCGUGGGACCA--AAAUGCCAAAGAUCAAGAA --------((((((...--...)))..((.(-(((((((((((((.............(((((((....)))))))))))))))))))))..)).--...)))............. ( -33.51, z-score = -4.86, R) >droSec1.super_7 201487 103 + 3727775 --------GCAUCUAAG--AUAAGACCGGGC-CACGCGGCGUAUGCGCAAUAAAACAAAUUAAUACAAAUAUUAAUCAUACGCCGCGUGGGACCA--AAAUGCCAAAGAUCAAGAA --------((((((...--...)))..((.(-(((((((((((((.............(((((((....)))))))))))))))))))))..)).--...)))............. ( -33.51, z-score = -4.86, R) >droYak2.chr2R 16590768 103 - 21139217 --------GCAUCUAAG--AUAAGACCAGGC-CACGCGGCGUAUGCGCAAUAAAACAAAUUAAUACAAAUAUUAAUUAUACGCCGCGUGGUAUGA--AAAUGUCAAGGAUCAAGAA --------(.((((..(--(((....((.((-(((((((((((((.............(((((((....)))))))))))))))))))))).)).--...))))..)))))..... ( -34.01, z-score = -5.39, R) >droEre2.scaffold_4845 19623690 103 + 22589142 --------GCAUCUAAG--AUAAGGCCGGGC-CACGCGGCGUAUGCGCAAUAAAACAAAUUAAUACAAAUAUUAAUCAUACGCCGCGUGGUACGA--AAAUGCCAAAGAUCAAGAA --------........(--((..(((((..(-(((((((((((((.............(((((((....)))))))))))))))))))))..)).--....)))....)))..... ( -36.81, z-score = -5.31, R) >droAna3.scaffold_12916 175835 103 - 16180835 --------ACAUCUCAG--AUAAGCUCGGAC-CACGCGGCGUAUGAGCAAUAAAACAAAUUAAUACUUAUAUUAAUCAUACGCCGCGUGGGACAC--AAAUGCCAAAGAUCAAGAA --------...(((..(--((..((..(..(-((((((((((((((..((((.................))))..)))))))))))))))..)..--....)).....))).))). ( -33.83, z-score = -5.98, R) >dp4.chr4_group3 8284209 100 + 11692001 -------------UUGG--AUAAGCCCCGGCGUAUGCGGUGUGUGCGCCAUAAAACAAAUUAAUACAGAUAUUAAUCAUACGCCGCGUGGGCCACC-AAAGGCCAAAGAUCAAGAG -------------((((--....(((((((((((((.((((....)))).........(((((((....))))))))))))))))...))))..))-))................. ( -31.30, z-score = -2.24, R) >droPer1.super_1 9733073 100 + 10282868 -------------UUGG--AUAAGCCCCGGCGUAUGCGGUGUGUGCGCCAUAAAACAAAUUAAUACAGAUAUUAAUCAUACGCUGCGUGGGCCACC-AAAGGCCAAAGAUCAAGAG -------------((((--....(((((((((((((.((((....)))).........(((((((....))))))))))))))))...))))..))-))................. ( -29.10, z-score = -1.53, R) >droGri2.scaffold_14978 740453 116 + 1124632 CCCCCACAGCAGAUAAACCAUAAGUUUGCGCUCAUGUGGCGUAUGCGCAAUAAAAUAAACUUACACACAUAUUAAUCAUACGCCGCUUGAGCCCAGUGAACAUUAAUGGGCAAAAA ..((((..((((((.........))))))(((((.((((((((((...((((.................))))...)))))))))).)))))..............))))...... ( -33.43, z-score = -3.62, R) >consensus ________GCAUCUAAG__AUAAGACCGGGC_CACGCGGCGUAUGCGCAAUAAAACAAAUUAAUACAAAUAUUAAUCAUACGCCGCGUGGGACCA__AAAUGCCAAAGAUCAAGAA ................................(((((((((((((.............(((((((....))))))))))))))))))))........................... (-21.53 = -21.13 + -0.40)

| Location | 16,554,817 – 16,554,920 |
|---|---|
| Length | 103 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 80.88 |
| Shannon entropy | 0.37926 |
| G+C content | 0.42973 |
| Mean single sequence MFE | -34.06 |
| Consensus MFE | -15.75 |
| Energy contribution | -16.43 |
| Covariance contribution | 0.68 |
| Combinations/Pair | 1.21 |
| Mean z-score | -3.34 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990247 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16554817 103 - 23011544 UUCUUGAUCUUUGGCAUUU--UGGUCCCACGCGGCGUAUGAUUAAUAUUUGUAUUAAUUUGUUUUAUUGCGCAUACGACGCGUG-GCCCGGUCUUAU--CUUAGAUGC-------- ......((((..(..(..(--(((..(((((((.(((((((((((((....)))))))..((......)).)))))).))))))-).))))...)..--)..))))..-------- ( -29.90, z-score = -2.35, R) >droSim1.chr2L 16250746 103 - 22036055 UUCUUGAUCUUUGGCAUUU--UGGUCCCACGCGGCGUAUGAUUAAUAUUUGUAUUAAUUUGUUUUAUUGCGCAUACGCCGCGUG-GCCCGGUCUUAU--CUUAGAUGC-------- ......((((..(..(..(--(((..(((((((((((((((((((((....)))))))..((......)).)))))))))))))-).))))...)..--)..))))..-------- ( -37.00, z-score = -4.47, R) >droSec1.super_7 201487 103 - 3727775 UUCUUGAUCUUUGGCAUUU--UGGUCCCACGCGGCGUAUGAUUAAUAUUUGUAUUAAUUUGUUUUAUUGCGCAUACGCCGCGUG-GCCCGGUCUUAU--CUUAGAUGC-------- ......((((..(..(..(--(((..(((((((((((((((((((((....)))))))..((......)).)))))))))))))-).))))...)..--)..))))..-------- ( -37.00, z-score = -4.47, R) >droYak2.chr2R 16590768 103 + 21139217 UUCUUGAUCCUUGACAUUU--UCAUACCACGCGGCGUAUAAUUAAUAUUUGUAUUAAUUUGUUUUAUUGCGCAUACGCCGCGUG-GCCUGGUCUUAU--CUUAGAUGC-------- .(((.(((....(((....--.((..(((((((((((((((((((((....)))))))).((......))..))))))))))))-)..)))))..))--)..)))...-------- ( -31.70, z-score = -4.24, R) >droEre2.scaffold_4845 19623690 103 - 22589142 UUCUUGAUCUUUGGCAUUU--UCGUACCACGCGGCGUAUGAUUAAUAUUUGUAUUAAUUUGUUUUAUUGCGCAUACGCCGCGUG-GCCCGGCCUUAU--CUUAGAUGC-------- .(((.(((....(((....--.((..(((((((((((((((((((((....)))))))..((......)).)))))))))))))-)..)))))..))--)..)))...-------- ( -37.90, z-score = -4.82, R) >droAna3.scaffold_12916 175835 103 + 16180835 UUCUUGAUCUUUGGCAUUU--GUGUCCCACGCGGCGUAUGAUUAAUAUAAGUAUUAAUUUGUUUUAUUGCUCAUACGCCGCGUG-GUCCGAGCUUAU--CUGAGAUGU-------- ......(((((.(((....--.((..(((((((((((((((.((((((((((....))))))...)))).))))))))))))))-)..)).)))...--..)))))..-------- ( -39.10, z-score = -5.64, R) >dp4.chr4_group3 8284209 100 - 11692001 CUCUUGAUCUUUGGCCUUU-GGUGGCCCACGCGGCGUAUGAUUAAUAUCUGUAUUAAUUUGUUUUAUGGCGCACACACCGCAUACGCCGGGGCUUAU--CCAA------------- .................((-((.(((((...((((((((((((((((....))))))).........((.(....).)).))))))))))))))...--))))------------- ( -30.30, z-score = -1.43, R) >droPer1.super_1 9733073 100 - 10282868 CUCUUGAUCUUUGGCCUUU-GGUGGCCCACGCAGCGUAUGAUUAAUAUCUGUAUUAAUUUGUUUUAUGGCGCACACACCGCAUACGCCGGGGCUUAU--CCAA------------- .................((-((.(((((.((..((((((((((((((....))))))).........((.(....).)).))))))))))))))...--))))------------- ( -26.90, z-score = -0.65, R) >droGri2.scaffold_14978 740453 116 - 1124632 UUUUUGCCCAUUAAUGUUCACUGGGCUCAAGCGGCGUAUGAUUAAUAUGUGUGUAAGUUUAUUUUAUUGCGCAUACGCCACAUGAGCGCAAACUUAUGGUUUAUCUGCUGUGGGGG ..((((((((...........)))(((((.(.((((((((..(((((...(((......)))..)))))..)))))))).).))))))))))((((((((......)))))))).. ( -36.70, z-score = -2.00, R) >consensus UUCUUGAUCUUUGGCAUUU__UGGUCCCACGCGGCGUAUGAUUAAUAUUUGUAUUAAUUUGUUUUAUUGCGCAUACGCCGCGUG_GCCCGGUCUUAU__CUUAGAUGC________ ...........(((.((......)).))).(((((((((((((((((....)))))))..((......)).))))))))))................................... (-15.75 = -16.43 + 0.68)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:32 2011