
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,548,839 – 16,548,932 |
| Length | 93 |
| Max. P | 0.863492 |

| Location | 16,548,839 – 16,548,932 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 79.25 |
| Shannon entropy | 0.38176 |
| G+C content | 0.46781 |
| Mean single sequence MFE | -22.50 |
| Consensus MFE | -12.91 |
| Energy contribution | -13.20 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.863492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16548839 93 - 23011544 --------CGACAAUGUCGCGACGCUUCCUAUUCAAUUUGAAAACCAAAUGAAGCGUUGCCUC---CGCCAAGG---AGCAAUUUACUCAGUGCAUUGAAUUGACCA --------...((((((.((((((((((.......(((((.....)))))))))))))))(((---(.....))---)).............))))))......... ( -27.01, z-score = -3.25, R) >droVir3.scaffold_12963 7261355 89 + 20206255 --------UGACAUGGACGCGUCGUUUCCC--UCAAUUUGUAAGCCAAAUGAAGCGUCGCCUC---UGCCAAGUUUCCCCCUUUUGCAGUGUGCAUUGACCA----- --------.......(((((.((((((..(--(.........))..)))))).)))))(((.(---(((.(((........))).)))).).))........----- ( -19.90, z-score = -0.85, R) >droWil1.scaffold_181038 546634 99 - 637489 CCAUCGCGUCUCAUUUUUCCGUUGCUUCCUUUUCAAUUUGAAAACCAAAUGAAGCGUCGCCUUCUACGCCAAGU---ACCAAUUUGCAGUGCGCAUUGACCA----- .((..((((..(.......((.((((((.(((((.....)))))......)))))).))........((.((((---....)))))).).))))..))....----- ( -15.30, z-score = 0.01, R) >dp4.chr4_group3 8275484 88 - 11692001 --------CGACUUUGUCGCGACGCUUCCUAUUCAAUUUGAAAACCAAAUGAUGCGUCCCCUC---CGCCAAGU---GCCAAUUUGCUCUGUGCAUUGACCA----- --------.......(((..(((((.((.......(((((.....))))))).))))).....---.((..((.---((......)).))..))...)))..----- ( -15.71, z-score = -0.79, R) >droPer1.super_1 9724713 88 - 10282868 --------CGACUUUGUCGCGACGCUUCCUAUUCAAUUUGAAAACCAAAUGAUGCGUCCCCUC---CGCCAAGU---GCCAAUUUGCUCUGUGCAUUGACCA----- --------.......(((..(((((.((.......(((((.....))))))).))))).....---.((..((.---((......)).))..))...)))..----- ( -15.71, z-score = -0.79, R) >droAna3.scaffold_12916 170045 88 + 16180835 --------CGACAAUGUCGCGACGCUUCCUAUUCAAUUUGAAAACCAAAUGAAGCGUCGUCUC---CGCCAAGU---GGCAAUUUGGCUUGUGCAGUGACCA----- --------.......(((((((((((((.......(((((.....))))))))))))).....---.(((((((---....))))))).......)))))..----- ( -29.21, z-score = -2.85, R) >droEre2.scaffold_4845 19617675 93 - 22589142 --------CCACAAUGUCGCGACGCUUCCUAUUCAAUUUGAAAACCAAAUGAAGCGUUGCCUG---CGCCAAGG---AGCAAUUUACCCAGUGCAUUGAAUUGACCA --------...((((((.((((((((((.......(((((.....))))))))))))))).((---(.(....)---.)))...........))))))......... ( -25.21, z-score = -2.75, R) >droYak2.chr2R 16584799 92 + 21139217 ---------CACAAUGUCGCGACGCUUCCUAUUCAAUUUAAAAACCAAAUGAAGCGUUGUCUC---CGCCAAGG---AGCAAUUUACCCAGUGCAUUGAAUUGACCA ---------..(((((.(((((((((((......................))))))))(.(((---(.....))---)))..........))))))))......... ( -22.95, z-score = -2.82, R) >droSec1.super_7 195488 93 - 3727775 --------CGACAAUGUCGCGACGCUUCCUAUUCAAUUUGAAAACCAAAUGAAGCGUUGCCUC---CGCCAAGG---AGCAAUUUACUCAGUGCAUUGAAUUGACCA --------...((((((.((((((((((.......(((((.....)))))))))))))))(((---(.....))---)).............))))))......... ( -27.01, z-score = -3.25, R) >droSim1.chr2L 16244756 93 - 22036055 --------CGACAAUGUCGCGACGCUUCCUAUUCAAUUUGAAAACCAAAUGAAGCGUUGCCUC---CGCCAAGG---AGCAAUUUACUCAGUGCAUUGAAUUGACCA --------...((((((.((((((((((.......(((((.....)))))))))))))))(((---(.....))---)).............))))))......... ( -27.01, z-score = -3.25, R) >consensus ________CGACAAUGUCGCGACGCUUCCUAUUCAAUUUGAAAACCAAAUGAAGCGUCGCCUC___CGCCAAGG___AGCAAUUUACUCAGUGCAUUGAACA_____ ..................((((((((((.......(((((.....)))))))))))))))...................((((.(((...))).))))......... (-12.91 = -13.20 + 0.29)

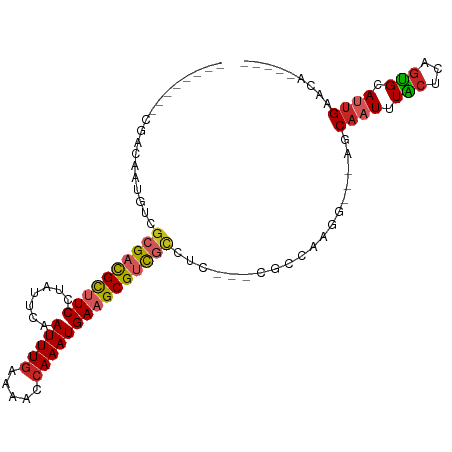
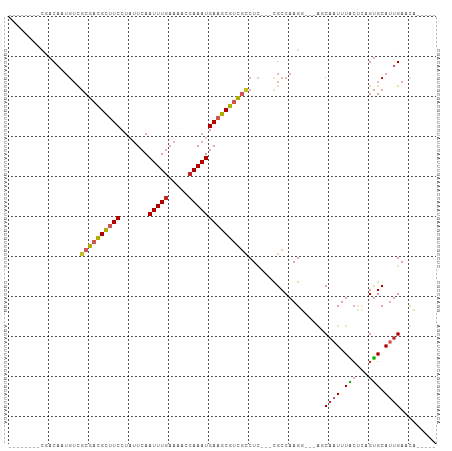
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:27 2011