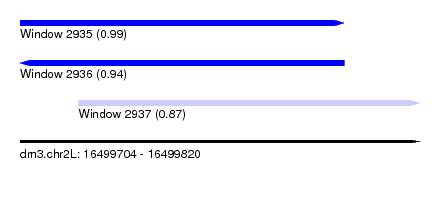
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,499,704 – 16,499,820 |
| Length | 116 |
| Max. P | 0.988607 |

| Location | 16,499,704 – 16,499,798 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 78.46 |
| Shannon entropy | 0.39616 |
| G+C content | 0.42880 |
| Mean single sequence MFE | -22.94 |
| Consensus MFE | -15.64 |
| Energy contribution | -15.77 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.33 |
| SVM RNA-class probability | 0.988607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
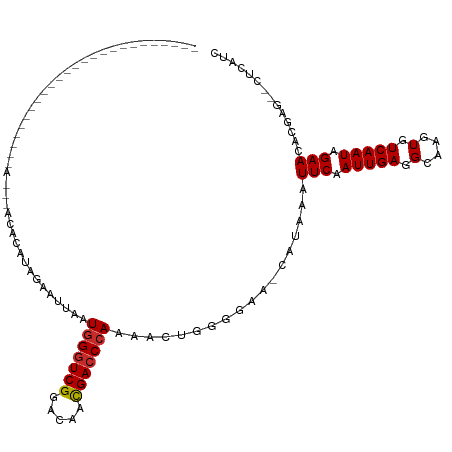
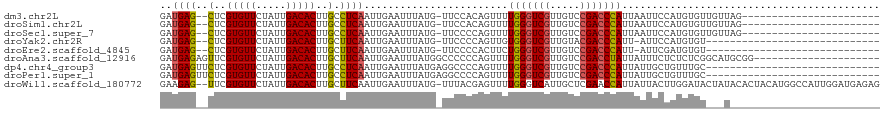
>dm3.chr2L 16499704 94 + 23011544 -----------------------CUAACAACACAUGGAAUUAAUGGGUCGGACAACGACCCAAAACUGUGGAA-CAUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAG--CUCAUC -----------------------.............(((((..(((((((.....)))))))....((((...-)))))))))...((((.(..(((((....).))))..)--)))).. ( -22.40, z-score = -2.26, R) >droSim1.chr2L 16209120 94 + 22036055 -----------------------CUAACAACACAUGGAAUUAAUGGGUCGGACAACGACCCAAAACUGUGGAA-CAUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAG--CUCAUC -----------------------.............(((((..(((((((.....)))))))....((((...-)))))))))...((((.(..(((((....).))))..)--)))).. ( -22.40, z-score = -2.26, R) >droSec1.super_7 153522 94 + 3727775 -----------------------CUAACAACACAUGGAAUUAAUGGGUCGGACAACGACCCAAAACUGGGGAA-CAUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAG--CUCAUC -----------------------.............(((((..(((((((.....)))))))......(....-)...)))))...((((.(..(((((....).))))..)--)))).. ( -23.20, z-score = -2.83, R) >droYak2.chr2R 16543422 87 - 21139217 -----------------------------ACACAUGGAAU-AAUGGGUCGUACAACGACCCACAACUGGGGAA-CAUAAAUUCAAUUGAAGCAAGUGUCAAUAGAACACGAG--CUCAUC -----------------------------.......((((-..(((((((.....)))))))......(....-)....))))...(((.((..(((((....).))))..)--)))).. ( -21.50, z-score = -2.43, R) >droEre2.scaffold_4845 19571186 87 + 22589142 -----------------------------ACACAUCGAAU-AAUGGGUCGGACAACGACCCAGAAGUGGGGAA-CAUAAAUUCAAUUGAAGCAAGUGUCAAUAGAACACGAG--CUCAUC -----------------------------.......((((-..(((((((.....)))))))...(((.....-)))..))))...(((.((..(((((....).))))..)--)))).. ( -23.40, z-score = -2.99, R) >droAna3.scaffold_12916 126937 99 - 16180835 ---------------------CCGCAUGCCGAGAGAGAAAUAAUAGGUCGGACAACGACCCAAAACUGGGGGGCCAUAAAUUCAAUUGAAGCAAGUGUCAAUAGAACACGAACUCUCAUC ---------------------.........(((((..........(((((.....)..((((....)))).)))).....(((.(((((........))))).)))......)))))... ( -22.20, z-score = -0.91, R) >dp4.chr4_group3 8227048 91 + 11692001 -----------------------------GCAAACAGCAAUAAUGGGUCGGACAACGACCCAAAACUGGGGCCUCAUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAGAACUCAUC -----------------------------((.....)).....(((((((.....)))))))....((((((((((..........))))))..(((((....).)))).....)))).. ( -29.70, z-score = -4.93, R) >droPer1.super_1 9675998 91 + 10282868 -----------------------------GCAAACAGCAAUAAUGGGUCGGACAACGACCCAAAACUGGGGCCUCAUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAGAACUCAUC -----------------------------((.....)).....(((((((.....)))))))....((((((((((..........))))))..(((((....).)))).....)))).. ( -29.70, z-score = -4.93, R) >droWil1.scaffold_180772 4771216 117 + 8906247 CUCUCAUCCAAUGGCCAUGUAGUGUAUAGUAUCCAAGUAAUAAUGGUUCGAGCAAUGACCCAAAACUCGUAAA-CAUAAAUUCAAUUGAAGCAAGUGUCAAUAGAACACGAA--CUCUUC ...........(((...((((...))))....))).........((((((....((((........))))...-......(((.(((((........))))).)))..))))--)).... ( -12.00, z-score = 1.92, R) >consensus _________________________A___ACACAUAGAAUUAAUGGGUCGGACAACGACCCAAAACUGGGGAA_CAUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAG__CUCAUC ...........................................(((((((.....)))))))..................(((.(((((.(....).))))).))).............. (-15.64 = -15.77 + 0.12)
| Location | 16,499,704 – 16,499,798 |
|---|---|
| Length | 94 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.46 |
| Shannon entropy | 0.39616 |
| G+C content | 0.42880 |
| Mean single sequence MFE | -23.17 |
| Consensus MFE | -15.66 |
| Energy contribution | -15.64 |
| Covariance contribution | -0.01 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.935362 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16499704 94 - 23011544 GAUGAG--CUCGUGUUCUAUUGACACUUGCCUCAAUUGAAUUUAUG-UUCCACAGUUUUGGGUCGUUGUCCGACCCAUUAAUUCCAUGUGUUGUUAG----------------------- ...(((--(....((((.(((((........))))).))))....)-)))((((.(..(((((((.....)))))))..)......)))).......----------------------- ( -23.10, z-score = -2.13, R) >droSim1.chr2L 16209120 94 - 22036055 GAUGAG--CUCGUGUUCUAUUGACACUUGCCUCAAUUGAAUUUAUG-UUCCACAGUUUUGGGUCGUUGUCCGACCCAUUAAUUCCAUGUGUUGUUAG----------------------- ...(((--(....((((.(((((........))))).))))....)-)))((((.(..(((((((.....)))))))..)......)))).......----------------------- ( -23.10, z-score = -2.13, R) >droSec1.super_7 153522 94 - 3727775 GAUGAG--CUCGUGUUCUAUUGACACUUGCCUCAAUUGAAUUUAUG-UUCCCCAGUUUUGGGUCGUUGUCCGACCCAUUAAUUCCAUGUGUUGUUAG----------------------- ...(((--(....((((.(((((........))))).))))....)-)))........(((((((.....))))))).((((..(....)..)))).----------------------- ( -21.10, z-score = -1.94, R) >droYak2.chr2R 16543422 87 + 21139217 GAUGAG--CUCGUGUUCUAUUGACACUUGCUUCAAUUGAAUUUAUG-UUCCCCAGUUGUGGGUCGUUGUACGACCCAUU-AUUCCAUGUGU----------------------------- ...(((--(..(((((.....)))))..))))((((((........-.....))))))(((((((.....)))))))..-...........----------------------------- ( -23.12, z-score = -2.41, R) >droEre2.scaffold_4845 19571186 87 - 22589142 GAUGAG--CUCGUGUUCUAUUGACACUUGCUUCAAUUGAAUUUAUG-UUCCCCACUUCUGGGUCGUUGUCCGACCCAUU-AUUCGAUGUGU----------------------------- ...(((--(....((((.(((((........))))).))))....)-)))..(((.(((((((((.....)))))))..-....)).))).----------------------------- ( -22.10, z-score = -2.35, R) >droAna3.scaffold_12916 126937 99 + 16180835 GAUGAGAGUUCGUGUUCUAUUGACACUUGCUUCAAUUGAAUUUAUGGCCCCCCAGUUUUGGGUCGUUGUCCGACCUAUUAUUUCUCUCUCGGCAUGCGG--------------------- ((.(((((.((((((((.(((((........))))).))))..))))......(((..(((((((.....)))))))..))).))))))).........--------------------- ( -22.80, z-score = -0.45, R) >dp4.chr4_group3 8227048 91 - 11692001 GAUGAGUUCUCGUGUUCUAUUGACACUUGCCUCAAUUGAAUUUAUGAGGCCCCAGUUUUGGGUCGUUGUCCGACCCAUUAUUGCUGUUUGC----------------------------- ...........(((((.....)))))..((((((..........))))))..((((..(((((((.....)))))))..))))........----------------------------- ( -26.70, z-score = -3.09, R) >droPer1.super_1 9675998 91 - 10282868 GAUGAGUUCUCGUGUUCUAUUGACACUUGCCUCAAUUGAAUUUAUGAGGCCCCAGUUUUGGGUCGUUGUCCGACCCAUUAUUGCUGUUUGC----------------------------- ...........(((((.....)))))..((((((..........))))))..((((..(((((((.....)))))))..))))........----------------------------- ( -26.70, z-score = -3.09, R) >droWil1.scaffold_180772 4771216 117 - 8906247 GAAGAG--UUCGUGUUCUAUUGACACUUGCUUCAAUUGAAUUUAUG-UUUACGAGUUUUGGGUCAUUGCUCGAACCAUUAUUACUUGGAUACUAUACACUACAUGGCCAUUGGAUGAGAG ((((..--...(((((.....)))))...)))).(((.((((((((-(...(((((..((...))..)))))..(((........)))............))))))..))).)))..... ( -19.80, z-score = 1.01, R) >consensus GAUGAG__CUCGUGUUCUAUUGACACUUGCCUCAAUUGAAUUUAUG_UUCCCCAGUUUUGGGUCGUUGUCCGACCCAUUAAUUCCAUGUGU___U_________________________ ..((((.....(((((.....)))))....))))........................(((((((.....)))))))........................................... (-15.66 = -15.64 + -0.01)
| Location | 16,499,721 – 16,499,820 |
|---|---|
| Length | 99 |
| Sequences | 8 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 85.82 |
| Shannon entropy | 0.27975 |
| G+C content | 0.40534 |
| Mean single sequence MFE | -26.61 |
| Consensus MFE | -15.56 |
| Energy contribution | -15.70 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.871620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
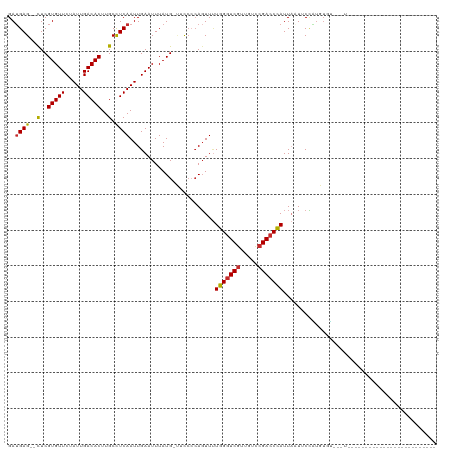
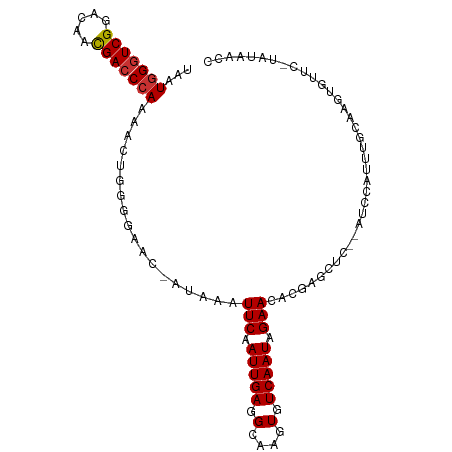
>dm3.chr2L 16499721 99 + 23011544 UAAUGGGUCGGACAACGACCCAAAACUGUGGAAC-AUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAGCUC--AUCCAUUUGUAAGUGUUC-UAUAACC ...(((((((.....)))))))....((((((((-((.....(((.((.(((..(((((....).))))..))).--...)).)))...))))))-))))... ( -30.80, z-score = -3.88, R) >droSim1.chr2L 16209137 99 + 22036055 UAAUGGGUCGGACAACGACCCAAAACUGUGGAAC-AUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAGCUC--AUCCAUUUGUAAGUGUUC-UAUAACC ...(((((((.....)))))))....((((((((-((.....(((.((.(((..(((((....).))))..))).--...)).)))...))))))-))))... ( -30.80, z-score = -3.88, R) >droSec1.super_7 153539 99 + 3727775 UAAUGGGUCGGACAACGACCCAAAACUGGGGAAC-AUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAGCUC--AUCCAUUUGUAAGUGUUC-UAUAACC ...(((((((.....))))))).......(((((-((.....(((.((.(((..(((((....).))))..))).--...)).)))...))))))-)...... ( -27.60, z-score = -2.64, R) >droYak2.chr2R 16543432 99 - 21139217 UAAUGGGUCGUACAACGACCCACAACUGGGGAAC-AUAAAUUCAAUUGAAGCAAGUGUCAAUAGAACACGAGCUC--AUCCAUUUUCAAGUGUUC-UAUAACC ...(((((((.....))))))).......(((((-((.........(((.((..(((((....).))))..))))--)...........))))))-)...... ( -25.95, z-score = -2.86, R) >droAna3.scaffold_12916 126956 102 - 16180835 UAAUAGGUCGGACAACGACCCAAAACUGGGGGGCCAUAAAUUCAAUUGAAGCAAGUGUCAAUAGAACACGAACUCUCAUCCAUAUGCAAGUUUUU-UAUAACU .....(((((.....)))))......((((((..(.....(((....)))....(((((....).)))))..))))))..........((((...-...)))) ( -19.20, z-score = -0.49, R) >dp4.chr4_group3 8227059 103 + 11692001 UAAUGGGUCGGACAACGACCCAAAACUGGGGCCUCAUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAGAACUCAUCCAUAUGCAAGCAUUCAUGUGACU ...(((((((.....)))))))....((((((((((..........))))))..(((((....).)))).....))))..((((((........))))))... ( -32.70, z-score = -3.60, R) >droPer1.super_1 9676009 103 + 10282868 UAAUGGGUCGGACAACGACCCAAAACUGGGGCCUCAUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAGAACUCAUCCAUAUGCAAGCAUUCAUGUGACU ...(((((((.....)))))))....((((((((((..........))))))..(((((....).)))).....))))..((((((........))))))... ( -32.70, z-score = -3.60, R) >droWil1.scaffold_180772 4771256 97 + 8906247 UAAUGGUUCGAGCAAUGACCCAAAACUCGUAAAC-AUAAAUUCAAUUGAAGCAAGUGUCAAUAGAACACGAACUCU--UCCAUUU-UACAUGUUG--AUGAUG ...(((.(((.....))).)))....((((.(((-((......(((.((((...(((((....).)))).....))--)).))).-...))))).--)))).. ( -13.10, z-score = 0.56, R) >consensus UAAUGGGUCGGACAACGACCCAAAACUGGGGAAC_AUAAAUUCAAUUGAGGCAAGUGUCAAUAGAACACGAGCUC__AUCCAUUUGCAAGUGUUC_UAUAACC ...(((((((.....)))))))..................(((.(((((.(....).))))).)))..................................... (-15.56 = -15.70 + 0.14)

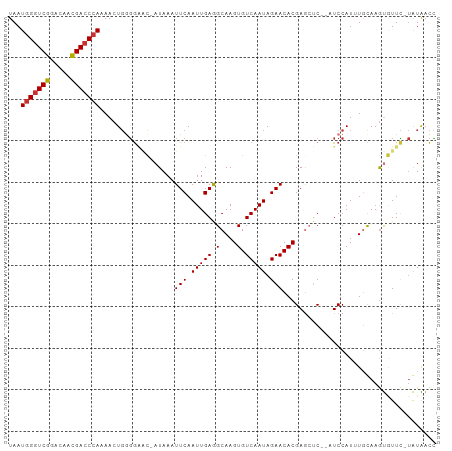
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:25 2011