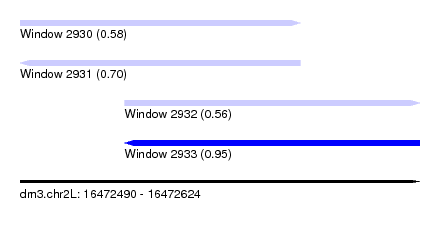
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 16,472,490 – 16,472,624 |
| Length | 134 |
| Max. P | 0.951016 |

| Location | 16,472,490 – 16,472,584 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 65.22 |
| Shannon entropy | 0.70479 |
| G+C content | 0.65841 |
| Mean single sequence MFE | -43.60 |
| Consensus MFE | -13.26 |
| Energy contribution | -14.01 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.53 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.576381 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
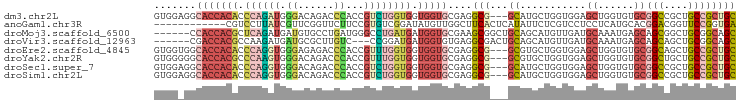
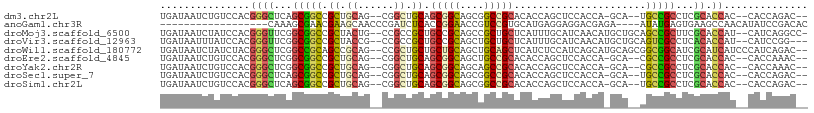
>dm3.chr2L 16472490 94 + 23011544 GUGGAGGCACCACACCCAGAUGGGACAGACCCACCGUCUGGUGGUGGUGCGAGGCG---GCAUGCUGGUGGAGCUGGUGUGCGGCCGCUGCCGCUGC (((((((((((((..((((((((..........))))))))..)))))))..(((.---((((((..(.....)..)))))).))).)).))))... ( -54.60, z-score = -3.18, R) >anoGam1.chr3R 3154225 85 + 53272125 ------------CGUCCUUAUCGUUCGGUUCUUCCGUGUCGGAUAUGUUGGCUUCACUCAUAUUCUCGUCCUCCUCAUGCACGGACGGUUCCGGUGA ------------............((((..(((((((((.(((((((..(......).)))))))..(.......)..))))))).))..))))... ( -20.30, z-score = -0.05, R) >droMoj3.scaffold_6500 7075959 91 + 32352404 ------CCACCACGCUCAGAUGAUGUGCCUGAUGGGCCUGAUGAUGGUGCGAAGCGGCUGCAGCAUGUUGAUGCAAAUGAGCAGCGGCUGCGGCAGC ------.(((((((.((((.......(((.....))))))))).)))))....((.((((((((.((((.((....)).))))...)))))))).)) ( -35.11, z-score = -0.23, R) >droVir3.scaffold_12963 7204784 88 - 20206255 ------CGACCACGCCAAGAUGAUGCGCUUGUC---CCGGAUGAUGGUGUGAGGCGACUGCAGCAUGUUGAUGCAAAUGAGCAGCAGCUGCGGCAGC ------....(((((((...(.((.((......---.)).)).))))))))..((..(((((((.(((((...(....)..))))))))))))..)) ( -30.50, z-score = -0.15, R) >droEre2.scaffold_4845 19539588 94 + 22589142 GUGGUGGCACCACACCCAGGUGGGAGAGACCCACCGUUUGGUGGUGGUGCGAGGCG---GCGUGCUGGUGGAGCUGGUGUGCGGCAGCUGCCGCUGC (..(((((((((((((..((((((.....))))))....))).)))))((...((.---(((..(..(.....)..)..))).)).)).)))))..) ( -52.00, z-score = -2.64, R) >droYak2.chr2R 2930796 94 + 21139217 GUGGGGGCACCACGCCCAAGUGGGACAGACCCACCGUUUGGUGGUGGUGCGAGGCG---GCGUGCUGGUGGAGCUGGUGUGCGGCUGCUGCCGCUGC ((((.((((((((..(((((((((.....)))))...))))..)))))....(((.---(((..(..(.....)..)..))).)))))).))))... ( -47.30, z-score = -0.82, R) >droSec1.super_7 118734 94 + 3727775 GUGGAGGCACCACACCCAGGUGGGACAGACCCACCGUCUGGUGGUGGUGCGAGGCG---GCAUGCUGGUGGAGCUGGUGUGCGGCCGCUGCCGCUGC ((((((((((((((((..((((((.....))))))....))).)))))))..(((.---((((((..(.....)..)))))).))).)).))))... ( -54.50, z-score = -2.59, R) >droSim1.chr2L 16182035 94 + 22036055 GUGGAGGCACCACACCCAGGUGGGACAGACCCACCGUCUGGUGGUGGUGCGAGGCG---GCAUGCUGGUGGAGCUGGUGUGCGGCCGCUGCCGCUGC ((((((((((((((((..((((((.....))))))....))).)))))))..(((.---((((((..(.....)..)))))).))).)).))))... ( -54.50, z-score = -2.59, R) >consensus GUGG_GGCACCACACCCAGAUGGGACAGACCCACCGUCUGGUGGUGGUGCGAGGCG___GCAUGCUGGUGGAGCUGGUGUGCGGCCGCUGCCGCUGC .......(((((((.(((((..((......))....))))))).)))))...............................(((((....)))))... (-13.26 = -14.01 + 0.75)
| Location | 16,472,490 – 16,472,584 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 65.22 |
| Shannon entropy | 0.70479 |
| G+C content | 0.65841 |
| Mean single sequence MFE | -35.46 |
| Consensus MFE | -14.62 |
| Energy contribution | -15.59 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.702475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16472490 94 - 23011544 GCAGCGGCAGCGGCCGCACACCAGCUCCACCAGCAUGC---CGCCUCGCACCACCACCAGACGGUGGGUCUGUCCCAUCUGGGUGUGGUGCCUCCAC ...((((((((((..((......))....)).)).)))---)))...(((((((..(((((...((((.....)))))))))..)))))))...... ( -42.00, z-score = -1.31, R) >anoGam1.chr3R 3154225 85 - 53272125 UCACCGGAACCGUCCGUGCAUGAGGAGGACGAGAAUAUGAGUGAAGCCAACAUAUCCGACACGGAAGAACCGAACGAUAAGGACG------------ ...(((((..(((((...........)))))....((((..((....)).)))))))....(((.....)))........))...------------ ( -16.50, z-score = 0.17, R) >droMoj3.scaffold_6500 7075959 91 - 32352404 GCUGCCGCAGCCGCUGCUCAUUUGCAUCAACAUGCUGCAGCCGCUUCGCACCAUCAUCAGGCCCAUCAGGCACAUCAUCUGAGCGUGGUGG------ .....((.(((.(((((......((((....)))).))))).))).))((((((..(((((((.....))).......))))..)))))).------ ( -30.11, z-score = 0.05, R) >droVir3.scaffold_12963 7204784 88 + 20206255 GCUGCCGCAGCUGCUGCUCAUUUGCAUCAACAUGCUGCAGUCGCCUCACACCAUCAUCCGG---GACAAGCGCAUCAUCUUGGCGUGGUCG------ (((((.(((..((.(((......))).))...))).)))))........(((((...((((---((...........)))))).)))))..------ ( -24.00, z-score = 0.98, R) >droEre2.scaffold_4845 19539588 94 - 22589142 GCAGCGGCAGCUGCCGCACACCAGCUCCACCAGCACGC---CGCCUCGCACCACCACCAAACGGUGGGUCUCUCCCACCUGGGUGUGGUGCCACCAC ...(((((.((((..((......)).....))))..))---)))...(((((((.(((....((((((.....))))))..))))))))))...... ( -43.10, z-score = -2.96, R) >droYak2.chr2R 2930796 94 - 21139217 GCAGCGGCAGCAGCCGCACACCAGCUCCACCAGCACGC---CGCCUCGCACCACCACCAAACGGUGGGUCUGUCCCACUUGGGCGUGGUGCCCCCAC ...(((((.((....))......(((.....)))..))---)))...(((((((..(((...((((((.....)))))))))..)))))))...... ( -39.80, z-score = -1.68, R) >droSec1.super_7 118734 94 - 3727775 GCAGCGGCAGCGGCCGCACACCAGCUCCACCAGCAUGC---CGCCUCGCACCACCACCAGACGGUGGGUCUGUCCCACCUGGGUGUGGUGCCUCCAC ...((((((((((..((......))....)).)).)))---)))...(((((((.(((....((((((.....))))))..))))))))))...... ( -44.10, z-score = -1.65, R) >droSim1.chr2L 16182035 94 - 22036055 GCAGCGGCAGCGGCCGCACACCAGCUCCACCAGCAUGC---CGCCUCGCACCACCACCAGACGGUGGGUCUGUCCCACCUGGGUGUGGUGCCUCCAC ...((((((((((..((......))....)).)).)))---)))...(((((((.(((....((((((.....))))))..))))))))))...... ( -44.10, z-score = -1.65, R) >consensus GCAGCGGCAGCGGCCGCACACCAGCUCCACCAGCAUGC___CGCCUCGCACCACCACCAGACGGUGGGUCUGUCCCACCUGGGCGUGGUGCC_CCAC ((.(((((....)))))......((...........))....))....((((((..((((..................))))..))))))....... (-14.62 = -15.59 + 0.97)
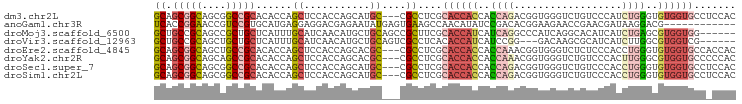
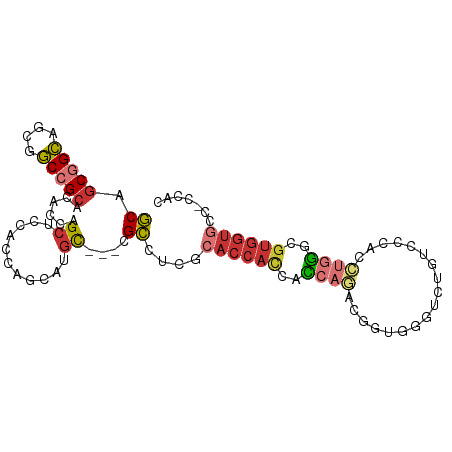
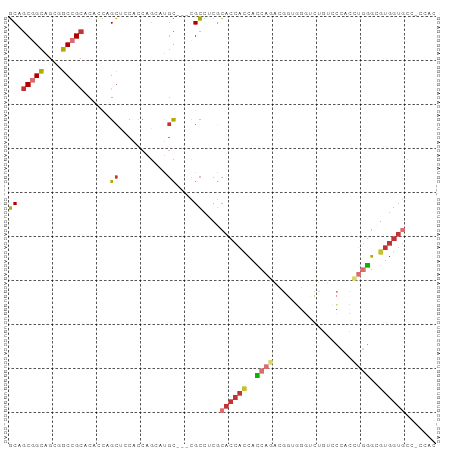
| Location | 16,472,525 – 16,472,624 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 70.76 |
| Shannon entropy | 0.60039 |
| G+C content | 0.65116 |
| Mean single sequence MFE | -45.83 |
| Consensus MFE | -22.72 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.53 |
| Mean z-score | -0.84 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.14 |
| SVM RNA-class probability | 0.563447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 16472525 99 + 23011544 --GUCUGGUG--GUGGUGCGAGGCGGCA--UGC-UGGUGGAGCUGGUGUGCGGCCGCUGCCGCUGCAGCCG--CUGCAGCGGCCGCUGAGCCCGUGGACAGAUUAUCA --(((((.(.--.(((.((..(((.(((--(((-..(.....)..)))))).)))((.((((((((((...--)))))))))).))...)))))..).)))))..... ( -53.70, z-score = -1.48, R) >anoGam1.chr3R 3154248 86 + 53272125 GUGUCGGAUAUGUUGGCUUCACUCAUAU----UCUCGUCCUCCUCAUGCACGGACGGUUCCGGUGAGAUCGGGUUGCUUCGUUCGCUUUG------------------ (((.((((((..(((..((((((.....----.(.(((((...........))))))....))))))..)))..)).))))..)))....------------------ ( -18.90, z-score = 1.29, R) >droMoj3.scaffold_6500 7075987 103 + 32352404 -GGCCUGAUG--AUGGUGCGAAGCGGCUGCAGCAUGUUGAUGCAAAUGAGCAGCGGCUGCGGCAGCGGCGG--CAGUAGCGGCCGCCGAACCCGUGGAUAGAUUAUCA -.....((((--((.((.((..((.((((((((.((((.((....)).))))...)))))))).))(((((--(.......)))))).......)).))..)))))). ( -43.40, z-score = -0.56, R) >droVir3.scaffold_12963 7204811 101 - 20206255 ---CCGGAUG--AUGGUGUGAGGCGACUGCAGCAUGUUGAUGCAAAUGAGCAGCAGCUGCGGCAGCGGCGG--CGGUAGCGGCCGCCGAACCCGUGGAUAAAUUAUCA ---.....(.--((((.((..((((.((((.(((.((((.(((......))).))))))).)))))(((.(--(....)).))))))..)))))).)........... ( -41.30, z-score = -1.19, R) >droWil1.scaffold_180772 4691366 104 + 8906247 --GUCUGAUGGGAUGAUGCGAUGCCGCCGCUGCAUGCUGAUGGAGAUGAGCUGCAGCUGCAGCAGCAGCGG--CUGCGGCUGCGGCCGAGCCCGUAGAUAGAUUAUCA --((((.(((((....(((..(((.((.((((((.(((.((....)).))))))))).)).)))))).(((--((((....)))))))..)))))))))......... ( -46.90, z-score = -1.30, R) >droEre2.scaffold_4845 19539623 99 + 22589142 --GUUUGGUG--GUGGUGCGAGGCGGCG--UGC-UGGUGGAGCUGGUGUGCGGCAGCUGCCGCUGCAGCCG--CUGCAGCGGCCGCCGAGCCCGUGGACAGAUUAUCA --((((((((--((.((....((((((.--(((-.((((((((((.(....).))))).))))))))))))--))...)).))))))))))................. ( -51.40, z-score = -0.96, R) >droYak2.chr2R 2930831 99 + 21139217 --GUUUGGUG--GUGGUGCGAGGCGGCG--UGC-UGGUGGAGCUGGUGUGCGGCUGCUGCCGCUGCAGCCG--CUGCAGCGGCCGCCGAGCCCGUGGACAGAUUAUCA --...(((((--((.(((((.(((((((--.((-((.((.(((.(.((((((((....))))).))).).)--)).)).))))))))..))))))..))..))))))) ( -49.50, z-score = -0.42, R) >droSec1.super_7 118769 99 + 3727775 --GUCUGGUG--GUGGUGCGAGGCGGCA--UGC-UGGUGGAGCUGGUGUGCGGCCGCUGCCGCUGCAGCCG--CUGCAGCGGCCGCUGAGCCCGUGGACAGAUUAUCA --(((((.(.--.(((.((..(((.(((--(((-..(.....)..)))))).)))((.((((((((((...--)))))))))).))...)))))..).)))))..... ( -53.70, z-score = -1.48, R) >droSim1.chr2L 16182070 99 + 22036055 --GUCUGGUG--GUGGUGCGAGGCGGCA--UGC-UGGUGGAGCUGGUGUGCGGCCGCUGCCGCUGCAGCCG--CUGCAGCGGCCGCUGAGCCCGUGGACAGAUUAUCA --(((((.(.--.(((.((..(((.(((--(((-..(.....)..)))))).)))((.((((((((((...--)))))))))).))...)))))..).)))))..... ( -53.70, z-score = -1.48, R) >consensus __GUCUGGUG__GUGGUGCGAGGCGGCA__UGC_UGGUGGAGCUGGUGUGCGGCCGCUGCCGCUGCAGCCG__CUGCAGCGGCCGCCGAGCCCGUGGACAGAUUAUCA ..............((.((..((((((......................(((((....))))).((.((......)).)).))))))..))))............... (-22.72 = -23.00 + 0.28)

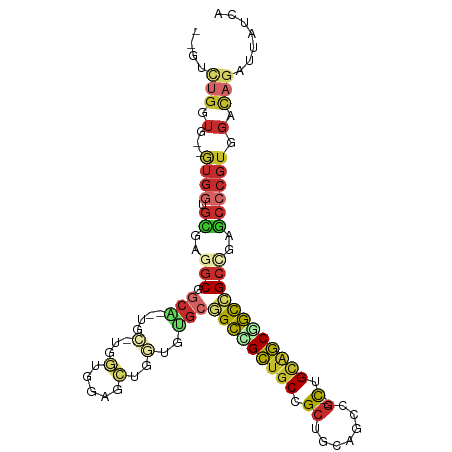
| Location | 16,472,525 – 16,472,624 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 108 |
| Reading direction | reverse |
| Mean pairwise identity | 70.76 |
| Shannon entropy | 0.60039 |
| G+C content | 0.65116 |
| Mean single sequence MFE | -36.96 |
| Consensus MFE | -21.91 |
| Energy contribution | -21.74 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951016 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
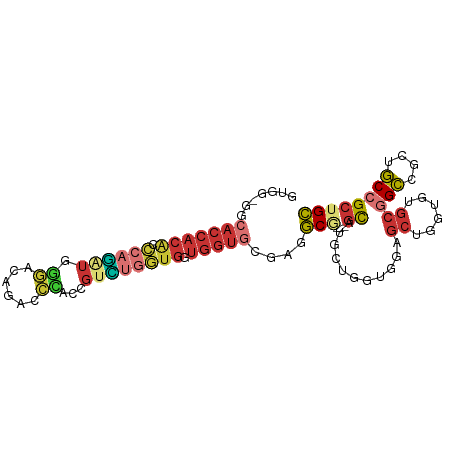
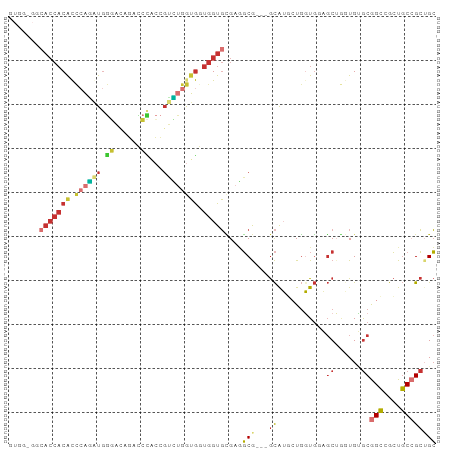
>dm3.chr2L 16472525 99 - 23011544 UGAUAAUCUGUCCACGGGCUCAGCGGCCGCUGCAG--CGGCUGCAGCGGCAGCGGCCGCACACCAGCUCCACCA-GCA--UGCCGCCUCGCACCAC--CACCAGAC-- ......((((.....((((...((((((((....)--))))))).((((((((((..((......))....)).-)).--))))))...)).))..--...)))).-- ( -40.60, z-score = -1.20, R) >anoGam1.chr3R 3154248 86 - 53272125 ------------------CAAAGCGAACGAAGCAACCCGAUCUCACCGGAACCGUCCGUGCAUGAGGAGGACGAGA----AUAUGAGUGAAGCCAACAUAUCCGACAC ------------------....((.......))...((..((((((((((....)))).)..))))).)).((.((----.((((..((....)).)))))))).... ( -14.90, z-score = 0.56, R) >droMoj3.scaffold_6500 7075987 103 - 32352404 UGAUAAUCUAUCCACGGGUUCGGCGGCCGCUACUG--CCGCCGCUGCCGCAGCCGCUGCUCAUUUGCAUCAACAUGCUGCAGCCGCUUCGCACCAU--CAUCAGGCC- ((((............(((.(((((((.......)--))))))..)))(((((.(((((......((((....)))).))))).)))..)).....--.))))....- ( -36.30, z-score = -0.76, R) >droVir3.scaffold_12963 7204811 101 + 20206255 UGAUAAUUUAUCCACGGGUUCGGCGGCCGCUACCG--CCGCCGCUGCCGCAGCUGCUGCUCAUUUGCAUCAACAUGCUGCAGUCGCCUCACACCAU--CAUCCGG--- ..............(((((..(((((......)))--))((.(((((.(((..((.(((......))).))...))).))))).))..........--.))))).--- ( -34.90, z-score = -1.61, R) >droWil1.scaffold_180772 4691366 104 - 8906247 UGAUAAUCUAUCUACGGGCUCGGCCGCAGCCGCAG--CCGCUGCUGCUGCAGCUGCAGCUCAUCUCCAUCAGCAUGCAGCGGCGGCAUCGCAUCAUCCCAUCAGAC-- ((((............((((((((....)))).))--))((((((((((((((((..............)))).)))))))))))).............))))...-- ( -43.84, z-score = -2.28, R) >droEre2.scaffold_4845 19539623 99 - 22589142 UGAUAAUCUGUCCACGGGCUCGGCGGCCGCUGCAG--CGGCUGCAGCGGCAGCUGCCGCACACCAGCUCCACCA-GCA--CGCCGCCUCGCACCAC--CACCAAAC-- ...............((((..((((((.((((..(--.(((((..(((((....)))))....))))).)..))-)).--.))))))..)).))..--........-- ( -40.60, z-score = -1.52, R) >droYak2.chr2R 2930831 99 - 21139217 UGAUAAUCUGUCCACGGGCUCGGCGGCCGCUGCAG--CGGCUGCAGCGGCAGCAGCCGCACACCAGCUCCACCA-GCA--CGCCGCCUCGCACCAC--CACCAAAC-- ...............((((..((((((.((((..(--.(((((..(((((....)))))....))))).)..))-)).--.))))))..)).))..--........-- ( -40.30, z-score = -1.62, R) >droSec1.super_7 118769 99 - 3727775 UGAUAAUCUGUCCACGGGCUCAGCGGCCGCUGCAG--CGGCUGCAGCGGCAGCGGCCGCACACCAGCUCCACCA-GCA--UGCCGCCUCGCACCAC--CACCAGAC-- ......((((.....((((...((((((((....)--))))))).((((((((((..((......))....)).-)).--))))))...)).))..--...)))).-- ( -40.60, z-score = -1.20, R) >droSim1.chr2L 16182070 99 - 22036055 UGAUAAUCUGUCCACGGGCUCAGCGGCCGCUGCAG--CGGCUGCAGCGGCAGCGGCCGCACACCAGCUCCACCA-GCA--UGCCGCCUCGCACCAC--CACCAGAC-- ......((((.....((((...((((((((....)--))))))).((((((((((..((......))....)).-)).--))))))...)).))..--...)))).-- ( -40.60, z-score = -1.20, R) >consensus UGAUAAUCUGUCCACGGGCUCGGCGGCCGCUGCAG__CGGCUGCAGCGGCAGCGGCCGCACACCAGCUCCACCA_GCA__AGCCGCCUCGCACCAC__CACCAGAC__ ...............((((...(((((.((.((......)).)).(((((....)))))......................)))))...)).)).............. (-21.91 = -21.74 + -0.17)
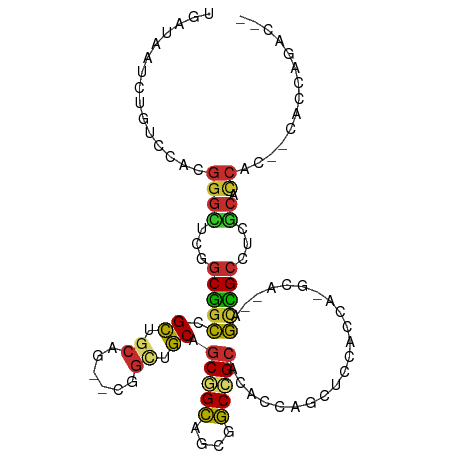
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:45:22 2011