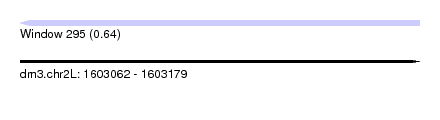
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,603,062 – 1,603,179 |
| Length | 117 |
| Max. P | 0.640390 |

| Location | 1,603,062 – 1,603,179 |
|---|---|
| Length | 117 |
| Sequences | 9 |
| Columns | 127 |
| Reading direction | reverse |
| Mean pairwise identity | 59.78 |
| Shannon entropy | 0.76511 |
| G+C content | 0.60830 |
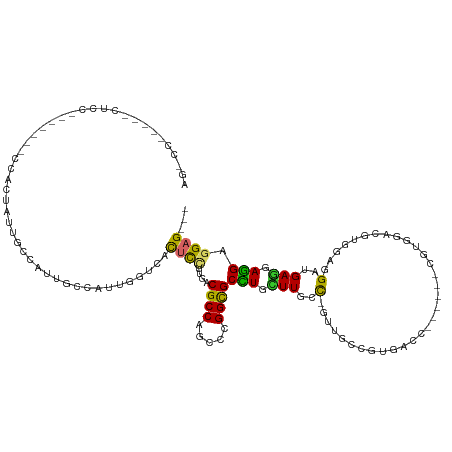
| Mean single sequence MFE | -41.63 |
| Consensus MFE | -10.24 |
| Energy contribution | -9.82 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.57 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.25 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.640390 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
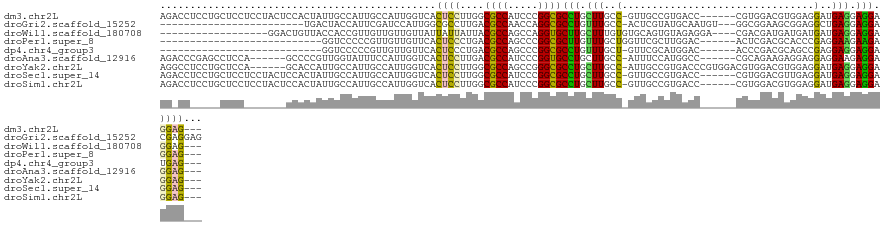
>dm3.chr2L 1603062 117 - 23011544 AGACCUCCUGCUCCUCCUACUCCACUAUUGCCAUUGCCAUUGGUCACUCCUUGGCGCCAUCCCGGCGCCUGCUUGCC-GUUGCCGUGACC------CGUGGACGUGGAGGAUGAGGAGGAGGAG--- ....(((((.((((((...((((((...........((((.((((((.....((((((.....)))))).((.....-...)).))))))------.))))..))))))...)))))).)))))--- ( -58.32, z-score = -5.22, R) >droGri2.scaffold_15252 10753746 99 - 17193109 ------------------------UGACUACCAUUCGAUCCAUUGGCGCCUUGACGCCAACCAGGCGCCUGUUUGCC-ACUCGUAUGCAAUGU---GGCGGAAGCGGAGGCUGAGGAGGACGAGGAG ------------------------......((..(((.(((.((((((......)))))).(((.(.((..((((((-((...........))---))))))...)).).)))....)))))))).. ( -34.70, z-score = -0.73, R) >droWil1.scaffold_180708 3408172 102 - 12563649 ------------------GGACUGUUACCACCGUUGUUGUUGUUAUUAUUAUUACGCCAGCCAGGUGCUUGCUUUGUGUGCAGUGUAGAGGA----CGACGAUGAUGAUGAUGAGGAGGAGGAG--- ------------------.........((.((.((.(..(..(((((((..((.((((.....)))).((((.(((....))).))))....----.))..)))))))..)..).)))).))..--- ( -23.80, z-score = 0.24, R) >droPer1.super_8 1410724 91 - 3966273 ---------------------------GGUCCCCCGUUGUUGUUCACUCCCUGACGCCAGCCCGGCGCUUGUUUGCUGGUUCGCUUGGAC------ACUCGACGCACCCGAGGAAGAAGAGGAG--- ---------------------------((....))...........((((((...((((((((((((......))))))...)).))).)------.((((.......)))).....)).))))--- ( -22.40, z-score = 2.33, R) >dp4.chr4_group3 10204153 90 - 11692001 ---------------------------GGUCCCCCGUUGUUGUUCACUCCCUGACGCCAGCCCGGCGCCUGUUUGCU-GUUCGCAUGGAC------ACCCGACGCAGCCGAGGAGGAGGAUGAG--- ---------------------------.((((.((((((.((((........)))).)))).((((...((((.(.(-((((....))))------).).))))..))))....)).))))...--- ( -26.40, z-score = 1.20, R) >droAna3.scaffold_12916 15002821 111 - 16180835 AGACCCGAGCCUCCA------GCCCCGUUGGUAUUUCCAUUGGUCACUCCUUGACGCCAUCCCGGUGCCUGCUUGCC-AUUUCCAUGGCC------CGCAGAAGAGGAGGAGGAAGAGGAGGAG--- ....((...((((..------.(((((.(((.....))).)))((.((((((..((((.....)))).((((..(((-((....))))).------.))))..))))))))))..)))).))..--- ( -41.20, z-score = -1.28, R) >droYak2.chr2L 1576238 117 - 22324452 AGGCCUCCUGCUCCA------GCACCAUUGCCAUUGCCAUUGGUCACUCCUUGGCGCCAGCCGGGCGCCUGCUUGCC-AUUGCCGUGACCCGUGGACGUGGACGUGGAGGAUGAGGAGGAGGAG--- ...((((((.(((..------.(.((((..((((..((((.((((((.....((((((.....)))))).((.....-...)).)))))).))))..))))..))))..)..))).))))))..--- ( -56.10, z-score = -2.32, R) >droSec1.super_14 1546142 117 - 2068291 AGACCUCCUGCUCCUCCUACUCCACUAUUGCCAUUGCCAUUGGUCACUCCUUGGCGCCAUCCCGGCGCCUGCUUGCC-GUUGCCGUGACC------CGUGGACGUUGAGGAUGAGGAGGAGGAG--- ...((((((.(((.(((((((((((....((....))....((((((.....((((((.....)))))).((.....-...)).))))))------.))))).))..)))).))).))))))..--- ( -53.40, z-score = -4.47, R) >droSim1.chr2L 1576470 117 - 22036055 AGACCUCCUGCUCCUCCUACUCCACUAUUGCCAUUGCCAUUGGUCACUCCUUGGCGCCAUCCCGGCGCCUGCUUGCC-GUUGCCGUGACC------CGUGGACGUGGAGGAUGAGGAGGAGGAG--- ....(((((.((((((...((((((...........((((.((((((.....((((((.....)))))).((.....-...)).))))))------.))))..))))))...)))))).)))))--- ( -58.32, z-score = -5.22, R) >consensus AG_CC_____CUCC_______CCACUAUUGCCAUUGCCAUUGGUCACUCCUUGACGCCAGCCCGGCGCCUGCUUGCC_GUUGCCGUGACC______CGUGGACGUGGAGGAUGAGGAGGAGGAG___ ..............................................((((....((((.....))))(((.(((......................................))).))).))))... (-10.24 = -9.82 + -0.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:45 2011